
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
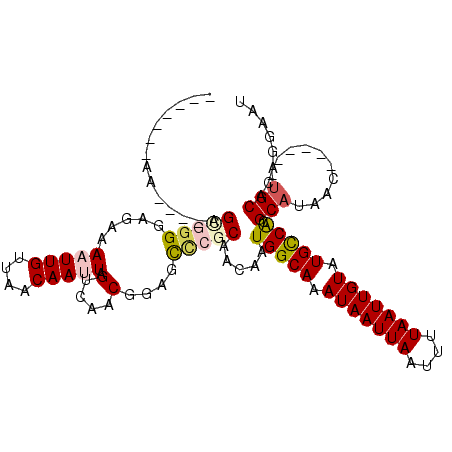
| Location | 8,649,524 – 8,649,632 |
| Length | 108 |
| Max. P | 0.925883 |

| Location | 8,649,524 – 8,649,632 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.71 |
| Mean single sequence MFE | -25.15 |
| Consensus MFE | -13.25 |
| Energy contribution | -13.83 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.831023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8649524 108 + 27905053 AUUGCAAAAAUUGAGCGGGGAGAAAAUUGUUAACAAUUAGUCAACGCUGUUCGCAACAAUGGCAAAUAAUUAAUUUUAAUUGUAUGCCAUGCAUAAC------AGCACAGGAAU ..(((.......(((((((..((.(((((....)))))..))..).))))))(((....(((((.(((((((....))))))).)))))))).....------.)))....... ( -27.50) >DroPse_CAF1 87471 103 + 1 -------AA----AGGGGGGGAAAAUUUGUUAACAAUUAGUCAACUGAGCCCGCAACAAUGGCAAAUAAUUAAUUUUAAUUGUAUGCCACGCAUAGCUGCAUCUGCAUAGGAAU -------..----.((((..((....(((....)))....))..)....)))(((....(((((.(((((((....))))))).))))).(((....)))...)))........ ( -22.60) >DroSec_CAF1 76382 108 + 1 AUUGCAAAAAUUGAGUGGGGAGAAAAUUGUUAACAAUUAGUCAACGCUAUUCGCAACAAUGGCAAAUAAUUAAUUUUAAUUGUAUGCCAUGCAUAAC------UGCACAGGAAU ..((((......(((((((..((.(((((....)))))..))..).))))))(((....(((((.(((((((....))))))).)))))))).....------))))....... ( -26.60) >DroEre_CAF1 89518 108 + 1 AUUGCAAAAAUUGGGCUGGGAGAAAAUUGUUAACAAUUAGUCAACGGUGCUCGCAACAAUGGCAAAUAAUUAAUUUUAAUUGUAUGUCGUGCAUAAC------UGCACAGGAAU .........((((...((.(((...((((((.((.....)).)))))).))).)).))))((((.(((((((....))))))).))))(((((....------)))))...... ( -26.10) >DroAna_CAF1 71205 95 + 1 -------------AGCGGGGAGAAAAUUGUUAACAAUUAGUCAACUGAGCCAGCAACAAUGGCAAAUAAUUAAUUUUAAUUGUAUGCCACGCAUAAG------UGCGCUGAAAC -------------.((.((..((.(((((....)))))..))..))..))((((.....(((((.(((((((....))))))).))))).(((....------))))))).... ( -25.50) >DroPer_CAF1 83586 102 + 1 -------AG----AGGG-GGGAAAAUUUGUUAACAAUUAGUCAACUGAGCCCGCAACAAUGGCAAAUAAUUAAUUUUAAUUGUAUGCCACGCAUAGAUGCAUCUGCAUAGGAAU -------..----.(((-........(((....)))((((....)))).)))(((....(((((.(((((((....))))))).))))).(((....)))...)))........ ( -22.60) >consensus _______AA____AGCGGGGAGAAAAUUGUUAACAAUUAGUCAACGGAGCCCGCAACAAUGGCAAAUAAUUAAUUUUAAUUGUAUGCCACGCAUAAC______UGCACAGGAAU ..............(((((.....(((((....))))).(....)....))))).....(((((.(((((((....))))))).))))).(((..........)))........ (-13.25 = -13.83 + 0.59)

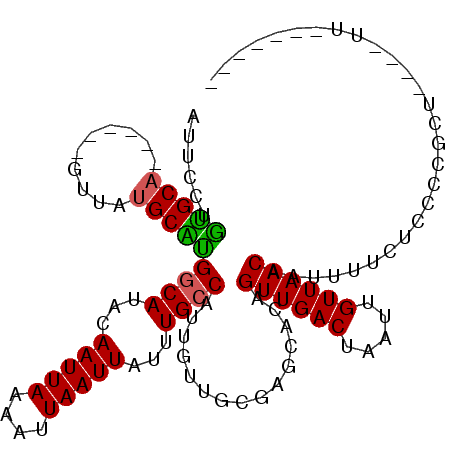
| Location | 8,649,524 – 8,649,632 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.71 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -15.28 |
| Energy contribution | -14.87 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8649524 108 - 27905053 AUUCCUGUGCU------GUUAUGCAUGGCAUACAAUUAAAAUUAAUUAUUUGCCAUUGUUGCGAACAGCGUUGACUAAUUGUUAACAAUUUUCUCCCCGCUCAAUUUUUGCAAU ........(((------(((.(((((((((...(((((....)))))...)))))....))))))))))((((((.....))))))............((.........))... ( -26.30) >DroPse_CAF1 87471 103 - 1 AUUCCUAUGCAGAUGCAGCUAUGCGUGGCAUACAAUUAAAAUUAAUUAUUUGCCAUUGUUGCGGGCUCAGUUGACUAAUUGUUAACAAAUUUUCCCCCCCU----UU------- ........((...((((((.....((((((...(((((....)))))...)))))).)))))).))...((((((.....))))))...............----..------- ( -23.80) >DroSec_CAF1 76382 108 - 1 AUUCCUGUGCA------GUUAUGCAUGGCAUACAAUUAAAAUUAAUUAUUUGCCAUUGUUGCGAAUAGCGUUGACUAAUUGUUAACAAUUUUCUCCCCACUCAAUUUUUGCAAU .......((((------(((((((((((((...(((((....)))))...)))))....)))..)))))((((((.....))))))......................)))).. ( -22.10) >DroEre_CAF1 89518 108 - 1 AUUCCUGUGCA------GUUAUGCACGACAUACAAUUAAAAUUAAUUAUUUGCCAUUGUUGCGAGCACCGUUGACUAAUUGUUAACAAUUUUCUCCCAGCCCAAUUUUUGCAAU .......((((------(...(((.((.((.(((((..((((.....))))...))))))))).)))..((((((.....)))))).....................))))).. ( -18.80) >DroAna_CAF1 71205 95 - 1 GUUUCAGCGCA------CUUAUGCGUGGCAUACAAUUAAAAUUAAUUAUUUGCCAUUGUUGCUGGCUCAGUUGACUAAUUGUUAACAAUUUUCUCCCCGCU------------- (..((((((((------....)))((((((...(((((....)))))...))))))....)))))..).((((((.....))))))...............------------- ( -23.40) >DroPer_CAF1 83586 102 - 1 AUUCCUAUGCAGAUGCAUCUAUGCGUGGCAUACAAUUAAAAUUAAUUAUUUGCCAUUGUUGCGGGCUCAGUUGACUAAUUGUUAACAAAUUUUCCC-CCCU----CU------- ........((((..(((....)))((((((...(((((....)))))...))))))..))))(((....((((((.....)))))).......)))-....----..------- ( -21.60) >consensus AUUCCUGUGCA______GUUAUGCAUGGCAUACAAUUAAAAUUAAUUAUUUGCCAUUGUUGCGAGCACAGUUGACUAAUUGUUAACAAUUUUCUCCCCGCU____UU_______ ......(((((..........)))))((((...(((((....)))))...))))...............((((((.....))))))............................ (-15.28 = -14.87 + -0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:24 2006