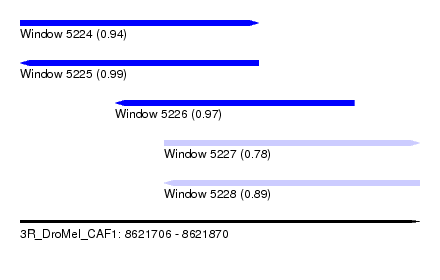
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,621,706 – 8,621,870 |
| Length | 164 |
| Max. P | 0.991481 |

| Location | 8,621,706 – 8,621,804 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -30.69 |
| Energy contribution | -31.17 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8621706 98 + 27905053 UGGCUUUUGCGGCGUUGUCACAGCCGAUGUGUCGUGCUGCAUGUUGCACACUGCGUUGCCACAUGUGUAGCACACUCGCAUUAACCAUGGAAAAGCGA ..((((((.((((.........)))((((((..(((((((((((.(((........))).))))))..)))))...))))))......).)))))).. ( -34.80) >DroSec_CAF1 49034 98 + 1 UGGCUUUUGCGGCGUUGUCACAGCCGAUGUGUCGUGCUGCAUGUUGCACACUGCGUUGCCACAUGUGUAGCACGCUCGCAUUAACCAAGGAAAAGCGA ..((((((.((((.........)))((((((.((((((((((((.(((........))).))))))..))))))..))))))......).)))))).. ( -37.70) >DroSim_CAF1 63346 98 + 1 UGGCUUUUGCGGCGUUGUCACAGCCGAUGUGUCGUGCUGCAUGUUGCACACUGCGUUGCCACAUGUGUAGCACACUCGCAUUAACCAAGGAAAAGCGA ..((((((.((((.........)))((((((..(((((((((((.(((........))).))))))..)))))...))))))......).)))))).. ( -35.00) >DroEre_CAF1 61148 98 + 1 UGGCUUUUGCGGCGUUGUCACAGCCGAUGUGUCGUGCUGCAUGUUGCACACUGCGUUGCCACAUGUGUAGCACACUCGCAUUAACCAAGGAGAAGCGA ..((((((.((((.........)))((((((..(((((((((((.(((........))).))))))..)))))...))))))......).)))))).. ( -35.10) >DroYak_CAF1 53411 98 + 1 UGGCUUUUGCGGCGUUGUCACAGCCGAUGUGUCGUGCUGCAUGUUGCACACUGCGUUGCCACAUGUGUAGCACACUCGCAUUAACCAAGGAAAAGCGA ..((((((.((((.........)))((((((..(((((((((((.(((........))).))))))..)))))...))))))......).)))))).. ( -35.00) >DroAna_CAF1 44954 96 + 1 CGGCUUUUGCGGCGUUGUCACAGCCGAUGUCUCGUGUUGCAUGUUGCACAUCCAUGUGCCACAUGUGCAA--CACUCACACAAGCAAAAAAAAAGAGG ....(((((((((((((.......)))))))..(((((((((((.(((((....))))).)))..)))))--)))........))))))......... ( -32.40) >consensus UGGCUUUUGCGGCGUUGUCACAGCCGAUGUGUCGUGCUGCAUGUUGCACACUGCGUUGCCACAUGUGUAGCACACUCGCAUUAACCAAGGAAAAGCGA ..((((((.((((.........)))((((((..(((((((((((.(((........))).)))..))))))))...))))))......).)))))).. (-30.69 = -31.17 + 0.47)

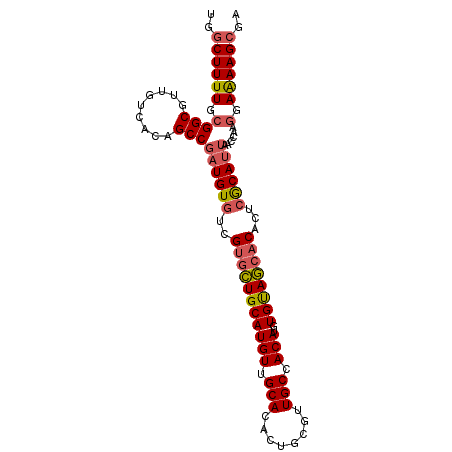
| Location | 8,621,706 – 8,621,804 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -37.38 |
| Consensus MFE | -31.78 |
| Energy contribution | -32.15 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8621706 98 - 27905053 UCGCUUUUCCAUGGUUAAUGCGAGUGUGCUACACAUGUGGCAACGCAGUGUGCAACAUGCAGCACGACACAUCGGCUGUGACAACGCCGCAAAAGCCA ...........(((((..((((.((((((.((((.((((....)))))))))).....(((((.(((....))))))))....))))))))..))))) ( -37.70) >DroSec_CAF1 49034 98 - 1 UCGCUUUUCCUUGGUUAAUGCGAGCGUGCUACACAUGUGGCAACGCAGUGUGCAACAUGCAGCACGACACAUCGGCUGUGACAACGCCGCAAAAGCCA ...........(((((..((((.((((((.((((.((((....)))))))))).....(((((.(((....))))))))....))))))))..))))) ( -39.50) >DroSim_CAF1 63346 98 - 1 UCGCUUUUCCUUGGUUAAUGCGAGUGUGCUACACAUGUGGCAACGCAGUGUGCAACAUGCAGCACGACACAUCGGCUGUGACAACGCCGCAAAAGCCA ...........(((((..((((.((((((.((((.((((....)))))))))).....(((((.(((....))))))))....))))))))..))))) ( -37.60) >DroEre_CAF1 61148 98 - 1 UCGCUUCUCCUUGGUUAAUGCGAGUGUGCUACACAUGUGGCAACGCAGUGUGCAACAUGCAGCACGACACAUCGGCUGUGACAACGCCGCAAAAGCCA ...........(((((..((((.((((((.((((.((((....)))))))))).....(((((.(((....))))))))....))))))))..))))) ( -37.60) >DroYak_CAF1 53411 98 - 1 UCGCUUUUCCUUGGUUAAUGCGAGUGUGCUACACAUGUGGCAACGCAGUGUGCAACAUGCAGCACGACACAUCGGCUGUGACAACGCCGCAAAAGCCA ...........(((((..((((.((((((.((((.((((....)))))))))).....(((((.(((....))))))))....))))))))..))))) ( -37.60) >DroAna_CAF1 44954 96 - 1 CCUCUUUUUUUUUGCUUGUGUGAGUG--UUGCACAUGUGGCACAUGGAUGUGCAACAUGCAACACGAGACAUCGGCUGUGACAACGCCGCAAAAGCCG ........(((((((((((((..(((--((((((((.((.....)).)))))))))))...))))))......(((.((....))))))))))))... ( -34.30) >consensus UCGCUUUUCCUUGGUUAAUGCGAGUGUGCUACACAUGUGGCAACGCAGUGUGCAACAUGCAGCACGACACAUCGGCUGUGACAACGCCGCAAAAGCCA ...........(((((..((((.((((((.((((.((((....)))))))))).....(((((.(((....))))))))....))))))))..))))) (-31.78 = -32.15 + 0.37)


| Location | 8,621,745 – 8,621,843 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 95.51 |
| Mean single sequence MFE | -30.95 |
| Consensus MFE | -29.40 |
| Energy contribution | -29.52 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8621745 98 - 27905053 ACAGCAUCCUGAGUUUUCCUGGGCUUUCCCGGGCCUUUCUCGCUUUUCCAUGGUUAAUGCGAGUGUGCUACACAUGUGGCAACGCAGUGUGCAACAUG ...(((..(((.(((..((((((....)))))).....(((((((..(....)..)).)))))...(((((....)))))))).)))..)))...... ( -32.80) >DroSec_CAF1 49073 98 - 1 ACAGCAUCCUGAGUUUUCCUGGGCUUUCCCGGGCCUUUCUCGCUUUUCCUUGGUUAAUGCGAGCGUGCUACACAUGUGGCAACGCAGUGUGCAACAUG ...(((..(((((....((((((....))))))...)))((((((..(....)..)).))))(((((((((....))))).))))))..)))...... ( -33.20) >DroSim_CAF1 63385 98 - 1 ACAGCAUCCUGAGUUUUCCUGGGCUUUCCCGGGCCUUUCUCGCUUUUCCUUGGUUAAUGCGAGUGUGCUACACAUGUGGCAACGCAGUGUGCAACAUG ...(((..(((.(((..((((((....)))))).....(((((((..(....)..)).)))))...(((((....)))))))).)))..)))...... ( -32.80) >DroEre_CAF1 61187 98 - 1 ACAGCAUCCUGAGUUUCCCUGAGUUUUCCCAAGCCUUUCUCGCUUCUCCUUGGUUAAUGCGAGUGUGCUACACAUGUGGCAACGCAGUGUGCAACAUG ...((((.(..((......((((...............))))......))..)...))))..((((((.((((.((((....)))))))))).)))). ( -24.06) >DroYak_CAF1 53450 98 - 1 ACAGCAUCCUGAGUUUUUCCCGGCUUUCCCGGGCCUUUCUCGCUUUUCCUUGGUUAAUGCGAGUGUGCUACACAUGUGGCAACGCAGUGUGCAACAUG ...(((..(((.(((...(((((.....))))).....(((((((..(....)..)).)))))...(((((....)))))))).)))..)))...... ( -31.90) >consensus ACAGCAUCCUGAGUUUUCCUGGGCUUUCCCGGGCCUUUCUCGCUUUUCCUUGGUUAAUGCGAGUGUGCUACACAUGUGGCAACGCAGUGUGCAACAUG ...(((..(((.(((..((((((....)))))).....(((((((..(....)..)).)))))...(((((....)))))))).)))..)))...... (-29.40 = -29.52 + 0.12)

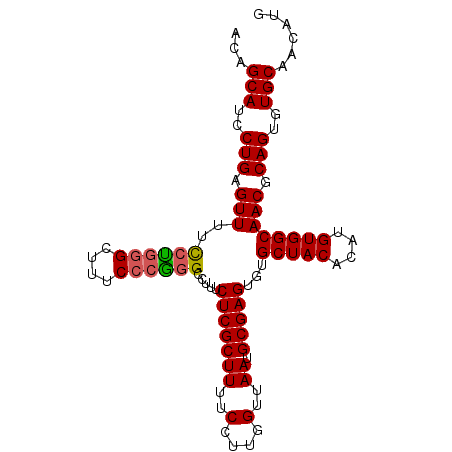
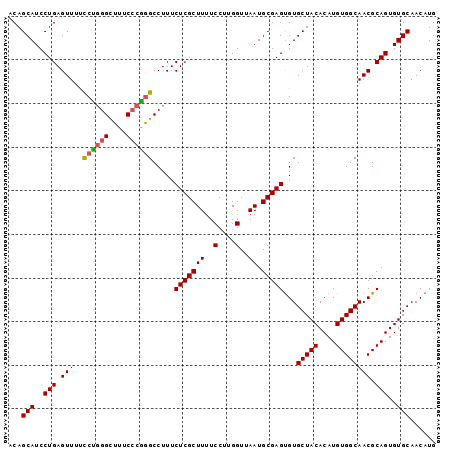
| Location | 8,621,765 – 8,621,870 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 93.14 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -24.56 |
| Energy contribution | -24.68 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.776893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8621765 105 + 27905053 CACAUGUGUAGCACACUCGCAUUAACCAUGGAAAAGCGAGAAAGGCCCGGGAAAGCCCAGGAAAACUCAGGAUGCUGUGAAUGGAGUGUGAAUGCGCGAAGCUGA ....((((((.(((((((.((((...(((((......(((.....((.(((....))).))....)))......))))))))))))))))..))))))....... ( -31.50) >DroSec_CAF1 49093 105 + 1 CACAUGUGUAGCACGCUCGCAUUAACCAAGGAAAAGCGAGAAAGGCCCGGGAAAGCCCAGGAAAACUCAGGAUGCUGUGAAUGGAGUGUGAAUGCGCGAAGCUGA ....((((((.(((((((.((((......((......(((.....((.(((....))).))....)))......))...)))))))))))..))))))....... ( -28.50) >DroSim_CAF1 63405 105 + 1 CACAUGUGUAGCACACUCGCAUUAACCAAGGAAAAGCGAGAAAGGCCCGGGAAAGCCCAGGAAAACUCAGGAUGCUGUGAAUGGAGUGUGAAUGCGCGAAGCUGA ....((((((.(((((((.((((......((......(((.....((.(((....))).))....)))......))...)))))))))))..))))))....... ( -28.90) >DroEre_CAF1 61207 99 + 1 CACAUGUGUAGCACACUCGCAUUAACCAAGGAGAAGCGAGAAAGGCUUGGGAAAACUCAGGGAAACUCAGGAUGCUGUGAAUGAAGUGUGAAUGCGCGA------ ....((((((.((((((((((.((.((...(((...(......).((((((....))))))....))).)).)).))))).....)))))..)))))).------ ( -24.50) >DroYak_CAF1 53470 105 + 1 CACAUGUGUAGCACACUCGCAUUAACCAAGGAAAAGCGAGAAAGGCCCGGGAAAGCCGGGAAAAACUCAGGAUGCUGUGAAUGGAGUGUGAAUGCGCGAAGCUGA ....((((((.(((((((.((((......((......(((.....(((((.....))))).....)))......))...)))))))))))..))))))....... ( -32.20) >consensus CACAUGUGUAGCACACUCGCAUUAACCAAGGAAAAGCGAGAAAGGCCCGGGAAAGCCCAGGAAAACUCAGGAUGCUGUGAAUGGAGUGUGAAUGCGCGAAGCUGA ....((((((.(((((((.((((...((..(......(((.....((.(((....))).))....)))......)..)))))))))))))..))))))....... (-24.56 = -24.68 + 0.12)


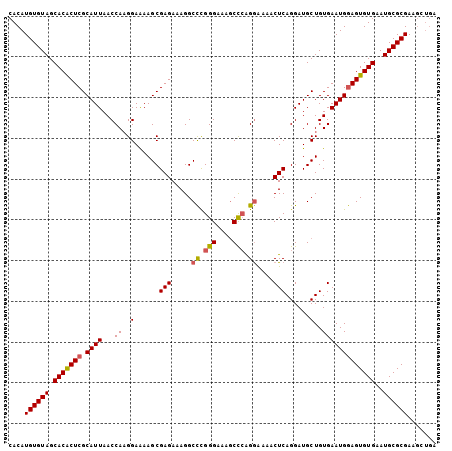
| Location | 8,621,765 – 8,621,870 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 93.14 |
| Mean single sequence MFE | -25.78 |
| Consensus MFE | -23.74 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.893071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8621765 105 - 27905053 UCAGCUUCGCGCAUUCACACUCCAUUCACAGCAUCCUGAGUUUUCCUGGGCUUUCCCGGGCCUUUCUCGCUUUUCCAUGGUUAAUGCGAGUGUGCUACACAUGUG ........((((((((.((..((((....(((.....(((....((((((....)))))).....)))))).....))))....)).)))))))).......... ( -29.10) >DroSec_CAF1 49093 105 - 1 UCAGCUUCGCGCAUUCACACUCCAUUCACAGCAUCCUGAGUUUUCCUGGGCUUUCCCGGGCCUUUCUCGCUUUUCCUUGGUUAAUGCGAGCGUGCUACACAUGUG ..(((..((((((((......(((.....(((.....(((....((((((....)))))).....))))))......)))..)))))..))).)))......... ( -24.70) >DroSim_CAF1 63405 105 - 1 UCAGCUUCGCGCAUUCACACUCCAUUCACAGCAUCCUGAGUUUUCCUGGGCUUUCCCGGGCCUUUCUCGCUUUUCCUUGGUUAAUGCGAGUGUGCUACACAUGUG ........((((((((.((..(((.....(((.....(((....((((((....)))))).....))))))......)))....)).)))))))).......... ( -28.50) >DroEre_CAF1 61207 99 - 1 ------UCGCGCAUUCACACUUCAUUCACAGCAUCCUGAGUUUCCCUGAGUUUUCCCAAGCCUUUCUCGCUUCUCCUUGGUUAAUGCGAGUGUGCUACACAUGUG ------..((((((((.(..........(((....))).(((..((.(((.......((((.......)))))))...))..)))).)))))))).......... ( -18.61) >DroYak_CAF1 53470 105 - 1 UCAGCUUCGCGCAUUCACACUCCAUUCACAGCAUCCUGAGUUUUUCCCGGCUUUCCCGGGCCUUUCUCGCUUUUCCUUGGUUAAUGCGAGUGUGCUACACAUGUG ........((((((((.((..(((.....(((.....(((.....(((((.....))))).....))))))......)))....)).)))))))).......... ( -28.00) >consensus UCAGCUUCGCGCAUUCACACUCCAUUCACAGCAUCCUGAGUUUUCCUGGGCUUUCCCGGGCCUUUCUCGCUUUUCCUUGGUUAAUGCGAGUGUGCUACACAUGUG ........((((((((.((..(((.....(((.....(((....((((((....)))))).....))))))......)))....)).)))))))).......... (-23.74 = -23.90 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:15 2006