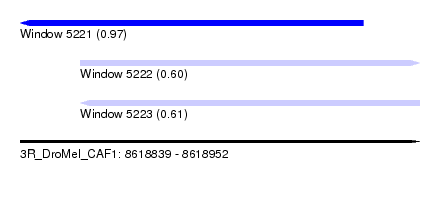
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,618,839 – 8,618,952 |
| Length | 113 |
| Max. P | 0.966682 |

| Location | 8,618,839 – 8,618,936 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.08 |
| Mean single sequence MFE | -20.28 |
| Consensus MFE | -12.96 |
| Energy contribution | -12.68 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
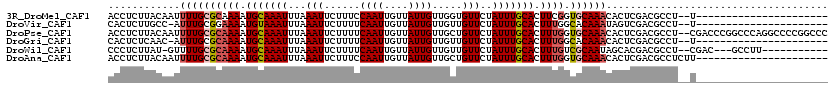
>3R_DroMel_CAF1 8618839 97 - 27905053 GCACCGAAGUGCAAAUAGAACACCAACAAUAACAAUUGGAAAGAAUUUAAAUUUGCAUUUUGCGCAAAAUUGUAAGAGGUA---UUC-----CGCCCCUCGUCU-U------- ((..((((((((((((..........((((....))))............)))))))))))).))..........((((..---...-----....))))....-.------- ( -18.35) >DroPse_CAF1 54771 96 - 1 GCACCAAAGUGCAAAUAGAACAGCAACAAUAACAAUUGAAAAGAAUUUAAAUUUGCAUUUUGCGCAAAAUUGUAAGAGGUU---CUC-----CUCUCCCAGCUC--------- ((..((((((((((((.(.....)..((((....))))............)))))))))))).))....(((..(((((..---..)-----))))..)))...--------- ( -21.80) >DroGri_CAF1 46760 106 - 1 GUGCCAAAGUGCAAAUAGAACAACAACAAUAACAAUUGAAAAGAAUUUAAAUUUGCAUUUUGCGCAAAU-GUUGAGAGUGAGACUAC-----CGUCUC-UAGAGGAGGGGGUG .(((((((((((((((..........((((....))))............)))))))))))).)))...-..............(((-----(.((((-.....)))).)))) ( -25.15) >DroWil_CAF1 70477 84 - 1 GCGACAAAGUGCAAAUAGAACAACAACAAUAACAAUUGAAAAGAAUUUAAAUUUGCAUUUUGCGCAAAAC-AUAAGAGGGC---UUC-----A-------------------- (((.((((((((((((..........((((....))))............))))))))))))))).....-..........---...-----.-------------------- ( -17.45) >DroAna_CAF1 42507 99 - 1 GCACCAAAGUGCAAAUAGAACAGCAACAAUAACAAUUGGAAAGAAUUUAAAUUUGCAUUUUGCGCAAAAUUGUAAGAGGUC---CUUUUUUUUCUCAC---UCG-C------- ((..((((((((((((..........((((....))))............)))))))))))).))......((.(((((..---......))))).))---...-.------- ( -17.15) >DroPer_CAF1 52465 96 - 1 GCACCAAAGUGCAAAUAGAACAGCAACAAUAACAAUUGAAAAGAAUUUAAAUUUGCAUUUUGCGCAAAAUUGUAAGAGGUU---CUC-----CUCUCCCAGCUC--------- ((..((((((((((((.(.....)..((((....))))............)))))))))))).))....(((..(((((..---..)-----))))..)))...--------- ( -21.80) >consensus GCACCAAAGUGCAAAUAGAACAGCAACAAUAACAAUUGAAAAGAAUUUAAAUUUGCAUUUUGCGCAAAAUUGUAAGAGGUA___CUC_____CGCCCC__GCCC_________ ((..((((((((((((..........((((....))))............)))))))))))).))................................................ (-12.96 = -12.68 + -0.28)
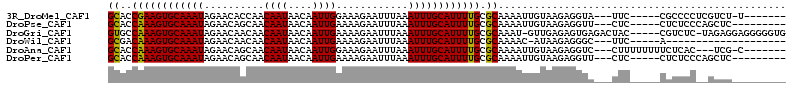
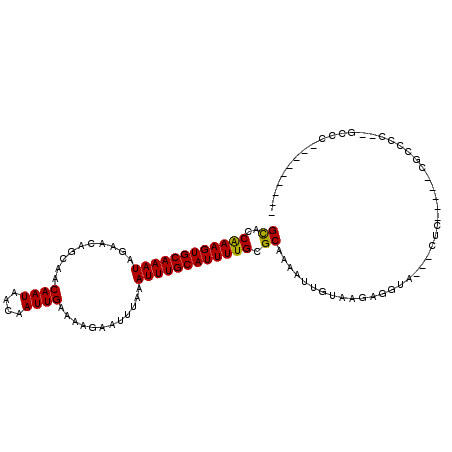
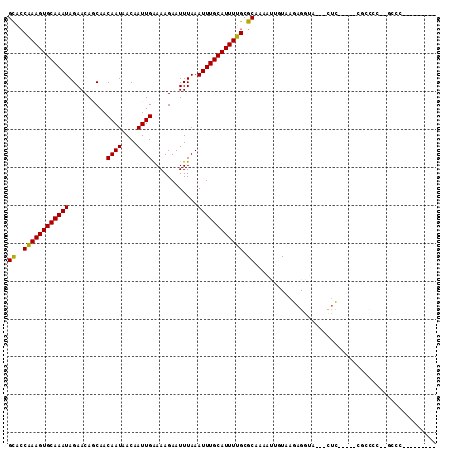
| Location | 8,618,856 – 8,618,952 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.83 |
| Mean single sequence MFE | -18.35 |
| Consensus MFE | -12.87 |
| Energy contribution | -13.32 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8618856 96 + 27905053 ACCUCUUACAAUUUUGCGCAAAAUGCAAAUUUAAAUUCUUUCCAAUUGUUAUUGUUGGUGUUCUAUUUGCACUUCGGUGCAAACACUCGACGCCU--U---------------------- ............((((((.....)))))).............((((....))))..((((((...(((((((....))))))).....)))))).--.---------------------- ( -21.70) >DroVir_CAF1 61599 95 + 1 CACUCUUGCC-AUUUGCGGAAAAUGUAAAUUUAAAUUCUUUUCAAUUGUUAUUGUUGUUGUUCUAUUUGCACUUUGGCACAAAUAGUCGACGCCU--U---------------------- .......((.-(((((((.....)))))))..........................((((..(((((((..(....)..))))))).))))))..--.---------------------- ( -12.70) >DroPse_CAF1 54787 118 + 1 ACCUCUUACAAUUUUGCGCAAAAUGCAAAUUUAAAUUCUUUUCAAUUGUUAUUGUUGCUGUUCUAUUUGCACUUUGGUGCAAACACUCGACGCCU--CGACCCGGCCCAGGCCCCGGCCC ............((((((((((.(((((((...(((......((((....)))).....)))..))))))).))).)))))))........(((.--......)))...(((....))). ( -24.80) >DroGri_CAF1 46787 95 + 1 CACUCUCAAC-AUUUGCGCAAAAUGCAAAUUUAAAUUCUUUUCAAUUGUUAUUGUUGUUGUUCUAUUUGCACUUUGGCACAAACACUCGACGCCU--U---------------------- ..........-...(((.((((.(((((((...(((......((((....)))).....)))..))))))).)))))))................--.---------------------- ( -14.10) >DroWil_CAF1 70482 103 + 1 CCCUCUUAU-GUUUUGCGCAAAAUGCAAAUUUAAAUUCUUUUCAAUUGUUAUUGUUGUUGUUCUAUUUGCACUUUGUCGCAAUAGCACGACGCCU--CGAC---GCCUU----------- ........(-((((((((((((.(((((((...(((......((((....)))).....)))..))))))).)))).))))).))))(((....)--))..---.....----------- ( -20.90) >DroAna_CAF1 42526 98 + 1 ACCUCUUACAAUUUUGCGCAAAAUGCAAAUUUAAAUUCUUUCCAAUUGUUAUUGUUGCUGUUCUAUUUGCACUUUGGUGCAAACACUCGACGCCUCUU---------------------- ............((((((((((.(((((((...(((...(..((((....))))..)..)))..))))))).))).)))))))...............---------------------- ( -15.90) >consensus ACCUCUUACAAUUUUGCGCAAAAUGCAAAUUUAAAUUCUUUUCAAUUGUUAUUGUUGUUGUUCUAUUUGCACUUUGGCGCAAACACUCGACGCCU__U______________________ ............((((((((((.(((((((...(((......((((....)))).....)))..))))))).)))).))))))..................................... (-12.87 = -13.32 + 0.44)
| Location | 8,618,856 – 8,618,952 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.83 |
| Mean single sequence MFE | -22.68 |
| Consensus MFE | -13.40 |
| Energy contribution | -14.15 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
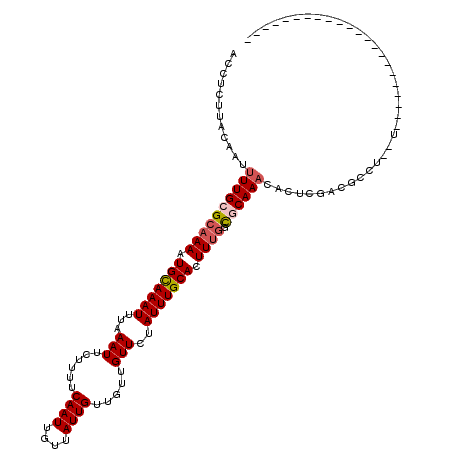
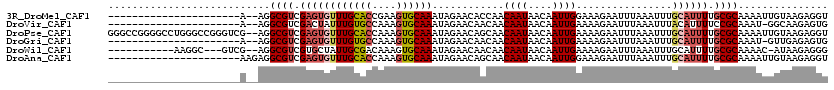
>3R_DroMel_CAF1 8618856 96 - 27905053 ----------------------A--AGGCGUCGAGUGUUUGCACCGAAGUGCAAAUAGAACACCAACAAUAACAAUUGGAAAGAAUUUAAAUUUGCAUUUUGCGCAAAAUUGUAAGAGGU ----------------------.--..((.((.....(((((..((((((((((((..........((((....))))............)))))))))))).))))).......)).)) ( -19.85) >DroVir_CAF1 61599 95 - 1 ----------------------A--AGGCGUCGACUAUUUGUGCCAAAGUGCAAAUAGAACAACAACAAUAACAAUUGAAAAGAAUUUAAAUUUACAUUUUCCGCAAAU-GGCAAGAGUG ----------------------.--..(((....(((((((..(....)..)))))))........((((....))))........................)))....-.......... ( -14.70) >DroPse_CAF1 54787 118 - 1 GGGCCGGGGCCUGGGCCGGGUCG--AGGCGUCGAGUGUUUGCACCAAAGUGCAAAUAGAACAGCAACAAUAACAAUUGAAAAGAAUUUAAAUUUGCAUUUUGCGCAAAAUUGUAAGAGGU ..(((((.((((.(((...))).--)))).))).)).(((((..((((((((((((.(.....)..((((....))))............)))))))))))).)))))............ ( -31.70) >DroGri_CAF1 46787 95 - 1 ----------------------A--AGGCGUCGAGUGUUUGUGCCAAAGUGCAAAUAGAACAACAACAAUAACAAUUGAAAAGAAUUUAAAUUUGCAUUUUGCGCAAAU-GUUGAGAGUG ----------------------.--..((.((((.(((((((((.(((((((((((..........((((....))))............)))))))))))))))))))-)))))..)). ( -23.05) >DroWil_CAF1 70482 103 - 1 -----------AAGGC---GUCG--AGGCGUCGUGCUAUUGCGACAAAGUGCAAAUAGAACAACAACAAUAACAAUUGAAAAGAAUUUAAAUUUGCAUUUUGCGCAAAAC-AUAAGAGGG -----------..(((---((..--..)))))(((...(((((.((((((((((((..........((((....))))............)))))))))))))))))..)-))....... ( -26.65) >DroAna_CAF1 42526 98 - 1 ----------------------AAGAGGCGUCGAGUGUUUGCACCAAAGUGCAAAUAGAACAGCAACAAUAACAAUUGGAAAGAAUUUAAAUUUGCAUUUUGCGCAAAAUUGUAAGAGGU ----------------------.....((.((.....(((((..((((((((((((..........((((....))))............)))))))))))).))))).......)).)) ( -20.15) >consensus ______________________A__AGGCGUCGAGUGUUUGCACCAAAGUGCAAAUAGAACAACAACAAUAACAAUUGAAAAGAAUUUAAAUUUGCAUUUUGCGCAAAAUUGUAAGAGGG ...........................((((.((((((((((((....))))))............((((....))))................)))))).))))............... (-13.40 = -14.15 + 0.75)
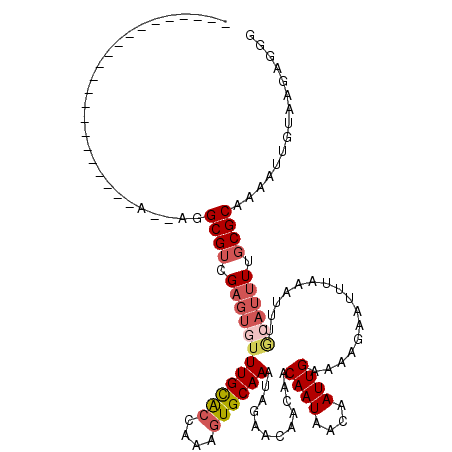

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:11 2006