| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,600,517 – 8,600,613 |
| Length | 96 |
| Max. P | 0.759633 |

| Location | 8,600,517 – 8,600,613 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.68 |
| Mean single sequence MFE | -24.68 |
| Consensus MFE | -11.51 |
| Energy contribution | -11.57 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8600517 96 - 27905053 GGCGU----GGCCCU-----CACAAUAGCCGGGGCCAUUGUCAUAAAUUAAAAAUGAGGAAGUGUAUGAAAAUAAACAAAAAGCCAAACGGCCC-----AGCCA----------GCUAGC (((((----((((((-----..(....)..))))))))...................((..((.(((....))).)).....(((....)))))-----.))).----------...... ( -26.20) >DroVir_CAF1 35033 103 - 1 GGCGU----GGCUGGCCGACAACAAUAGCCA-GGCCAUUGUCAUAAAUUAAAAAUGAGGAAGUAUAUGAAAUUUAACAAAAA--CAAACGACCC-GCUGAACAA---------CAGCAAC (((.(----(((((...........))))))-.))).((.((((.........)))).))......................--..........-((((.....---------))))... ( -22.70) >DroPse_CAF1 34789 101 - 1 -------CCGGCUUU-----AACAAAAGCCGGGGCCAUUGUCAUAAAUUAAAAAUGAGGAAGUGUAUGAAAAUAAACAAAAAGC-AAACAGCCGCAUGGAACCA--A----AGCUCCAGC -------((((((((-----....))))))))(((..((.((((.........)))).)).((.(((....))).)).......-.....)))((.((((....--.----...)))))) ( -26.10) >DroGri_CAF1 27218 112 - 1 GGCGU----GGCCAGCCGACAACAAUAGCCA-GGCCAUUGUCAUAAAUUAAAAAUGAGGAAGUAUAUGAAAUUUAACAAAAA--CAAACGGCCC-GCGGAACAGCACCAGCAGCAGCAGC .(((.----((((.(((..............-)))..((.((((.........)))).))......................--.....)))))-))((.......)).((....))... ( -21.14) >DroMoj_CAF1 57525 110 - 1 GGCGU----GGCUGGCCGACAACAAUAGCCA-GGCCAUUGUCAUAAAUUAAAAAUGAGGAAGUAUAUGAAAUUUAACAACAA--CAAACGGCCC-GCGGAACAA--ACAGCAGCAGCAGC .(((.----((((((((..............-)))).((.((((.........)))).))......................--.....)))))-)).......--...((....))... ( -21.94) >DroPer_CAF1 32393 108 - 1 UCCGUCCCCGGCUUU-----AACAAAAGCCGGGGCCAUUGUCAUAAAUUAAAAAUGAGGAAGUGUAUGAAAAUAAACAAAAAGC-AAACAGCCGCAUGGAACCA--A----AGCUCCAGC ((((.((((((((((-----....)))))))))).)....((((.........))))))).((.(((....))).)).....((-.....)).((.((((....--.----...)))))) ( -30.00) >consensus GGCGU____GGCUGG_____AACAAUAGCCA_GGCCAUUGUCAUAAAUUAAAAAUGAGGAAGUAUAUGAAAAUAAACAAAAA__CAAACGGCCC_GCGGAACAA__A____AGCACCAGC .........((((.........((((.((....)).))))(((((.(((...........))).)))))....................))))........................... (-11.51 = -11.57 + 0.06)

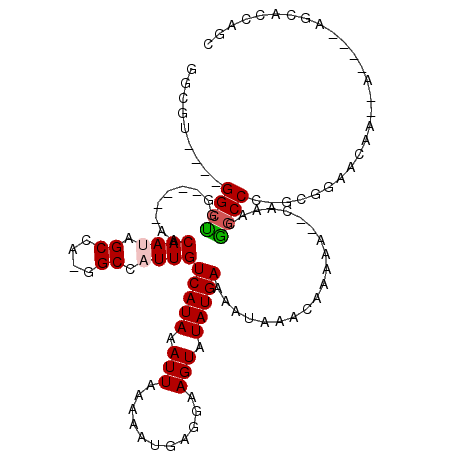

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:06 2006