
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
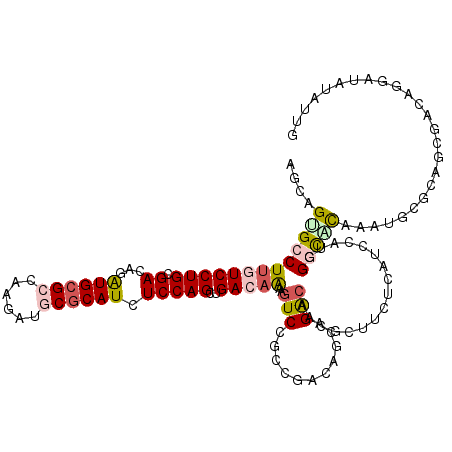
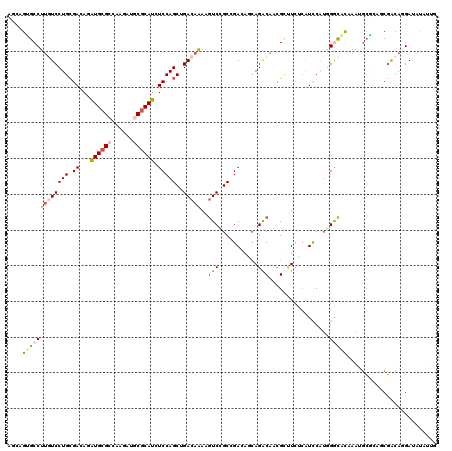
| Location | 8,532,107 – 8,532,227 |
| Length | 120 |
| Max. P | 0.836377 |

| Location | 8,532,107 – 8,532,227 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.17 |
| Mean single sequence MFE | -42.77 |
| Consensus MFE | -15.25 |
| Energy contribution | -15.75 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8532107 120 + 27905053 AACAGAGCCUUAUCCUGAGACAUGUGGGCAAGGAUCCGCAUCUCCAGCUGACAGAAGUCAGCCGACAAAAGACAUCGGCUCUCAUCCGUGGGCAUGAAUGGUGACCUACAGGAUAUGUGG .((((((((.......((((..((((((......))))))))))..((((((....))))))..............)))))..((((((((((((.....))).))))).)))).))).. ( -41.10) >DroVir_CAF1 17135 120 + 1 AGCAGCGCCUUGUCCUGCGACAGAUGCGCCAAGAUGCGCAUCUCCAGCUGACAAAAGUCUGCCGAAAGCAGGCAAUGCUUCUCGUUCGACGACACAAAUGCGCUGCGACAGGAUAUGUUA .(((((((.(((((..((...((((((((......))))))))...)).)))))..(((...((((((((.....))))).)))...))).........)))))))((((.....)))). ( -47.70) >DroGri_CAF1 17931 120 + 1 AGCAGUGCCUGAUCCUGCGACAGGUGCGCUAAAAUGCGCAUCUCCAGCUGACAAAAAUCGGCGGAGAGCAGACACCGUUUCCCAUCCAUUGGUGCAAAGGCGCUGCGACACGAUAUAUUG .(((((((((....((((......(((((......)))))(((((.(((((......)))))))))))))).(((((............)))))...))))))))).............. ( -50.30) >DroMoj_CAF1 11204 120 + 1 AGCAGUGCCUUGUCCUGCGAGAGAUGCGCCAAGAUGCGCAUCUCCAGCUGACAGAAGUCUGCAGAGAUCAGGCAACGCUUCUCAUCCAUGGGCGCAAAGGCGCUGCGACACGAGAUCUUC .((((((((((((((((.(((((((((((......)))))))))..((.(((....))).)).((((...(....)...)))).)))).))))...)))))))))).............. ( -52.90) >DroAna_CAF1 10023 120 + 1 ACAAGUGCCUUGUCCUGCGAUAAAUGCGCCAAAAUUCGCAUCUCCAGCUGACAAAAGUCCGCCGACAACAGGCAACGCGUAGAAUCCAUGGGCAUGAAUGCGGAUCGGCAAGAUACAAUG ....(((.(((((((((((....((((.(((.....(((..........(((....))).(((.......)))...))).........)))))))...)))))...)))))).))).... ( -31.34) >DroPer_CAF1 10425 120 + 1 ACCAGGGCCUUGUCCUGCGACAAAUGCGCCAAUAUGCGCAUCUCCAGCUGACAGAAGUCCGCCGAUAGCAGACAGCGCUUCUCGUCCAUGGACUUGAAUACUGAUCGACAGGAAAUACGA ........((((((((((.....((((((......)))))).....((.(((....))).)).....)))).(((...(((..(((....)))..)))..)))...))))))........ ( -33.30) >consensus AGCAGUGCCUUGUCCUGCGACAGAUGCGCCAAGAUGCGCAUCUCCAGCUGACAAAAGUCCGCCGACAGCAGACAACGCUUCUCAUCCAUGGGCACAAAUGCGCAGCGACAGGAUAUAUUG ....(((((((((((((.((...((((((......)))))).)))))..)))))..(((...........))).................)))))......................... (-15.25 = -15.75 + 0.50)

| Location | 8,532,107 – 8,532,227 |
|---|---|
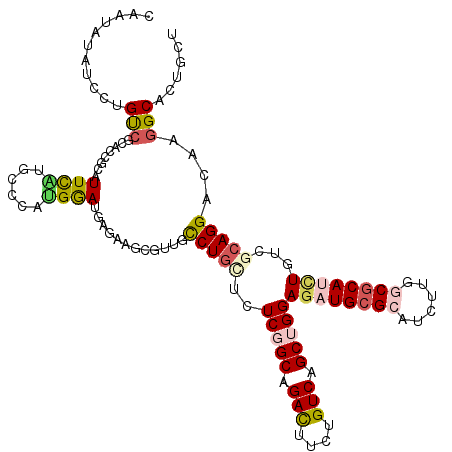
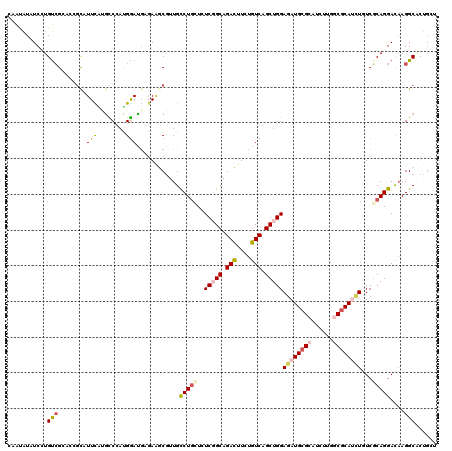
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.17 |
| Mean single sequence MFE | -45.00 |
| Consensus MFE | -21.77 |
| Energy contribution | -22.50 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8532107 120 - 27905053 CCACAUAUCCUGUAGGUCACCAUUCAUGCCCACGGAUGAGAGCCGAUGUCUUUUGUCGGCUGACUUCUGUCAGCUGGAGAUGCGGAUCCUUGCCCACAUGUCUCAGGAUAAGGCUCUGUU .....(((((.((.(((..........))).)))))))((((((..((((((....((((((((....))))))))((.(((.((........)).))).))..)))))).))))))... ( -42.60) >DroVir_CAF1 17135 120 - 1 UAACAUAUCCUGUCGCAGCGCAUUUGUGUCGUCGAACGAGAAGCAUUGCCUGCUUUCGGCAGACUUUUGUCAGCUGGAGAUGCGCAUCUUGGCGCAUCUGUCGCAGGACAAGGCGCUGCU ..............(((((((...(((.(((.....)))...)))((((((((..(((((.(((....))).)))))((((((((......))))))))...))))).))).))))))). ( -51.70) >DroGri_CAF1 17931 120 - 1 CAAUAUAUCGUGUCGCAGCGCCUUUGCACCAAUGGAUGGGAAACGGUGUCUGCUCUCCGCCGAUUUUUGUCAGCUGGAGAUGCGCAUUUUAGCGCACCUGUCGCAGGAUCAGGCACUGCU .........(((((((((((((.((...(((.....)))..)).)))).))))(((((((.(((....))).)).)))))(((((......)))))((((...))))....))))).... ( -43.40) >DroMoj_CAF1 11204 120 - 1 GAAGAUCUCGUGUCGCAGCGCCUUUGCGCCCAUGGAUGAGAAGCGUUGCCUGAUCUCUGCAGACUUCUGUCAGCUGGAGAUGCGCAUCUUGGCGCAUCUCUCGCAGGACAAGGCACUGCU .....((((((.(((..((((....))))...)))))))))((((.(((((..((.((((.(((....)))....((((((((((......)))))))))).))))))..))))).)))) ( -53.50) >DroAna_CAF1 10023 120 - 1 CAUUGUAUCUUGCCGAUCCGCAUUCAUGCCCAUGGAUUCUACGCGUUGCCUGUUGUCGGCGGACUUUUGUCAGCUGGAGAUGCGAAUUUUGGCGCAUUUAUCGCAGGACAAGGCACUUGU ..........((((....(((((((((....)))))).....)))((((((((..(((((.(((....))).)))))(((((((........)))))))...))))).)))))))..... ( -36.30) >DroPer_CAF1 10425 120 - 1 UCGUAUUUCCUGUCGAUCAGUAUUCAAGUCCAUGGACGAGAAGCGCUGUCUGCUAUCGGCGGACUUCUGUCAGCUGGAGAUGCGCAUAUUGGCGCAUUUGUCGCAGGACAAGGCCCUGGU ........((((((...((((.(((..(((....)))..)))..)))).((((..(((((.(((....))).)))))((((((((......))))))))...))))))).)))....... ( -42.50) >consensus CAAUAUAUCCUGUCGCACCGCAUUCAUGCCCAUGGAUGAGAAGCGUUGCCUGCUCUCGGCAGACUUCUGUCAGCUGGAGAUGCGCAUCUUGGCGCAUCUGUCGCAGGACAAGGCACUGCU ...........(((........((((......))))............(((((..(((((.(((....))).)))))((((((((......))))))))...)))))....)))...... (-21.77 = -22.50 + 0.73)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:42 2006