
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
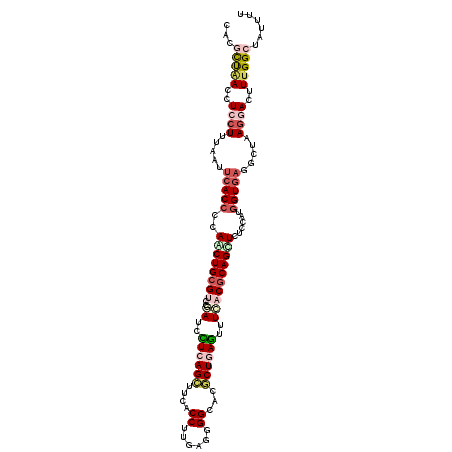
| Location | 8,531,395 – 8,531,509 |
| Length | 114 |
| Max. P | 0.898247 |

| Location | 8,531,395 – 8,531,509 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 82.92 |
| Mean single sequence MFE | -34.29 |
| Consensus MFE | -24.53 |
| Energy contribution | -26.37 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
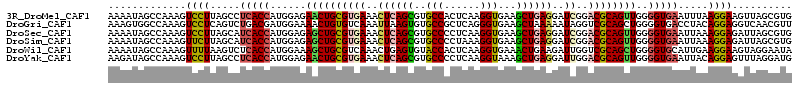
>3R_DroMel_CAF1 8531395 114 + 27905053 CACGCUAACUUCCUUAAAUUCACCCCAACUGCGUCCGAUCCUCAGCUUCACCUUGAGUGGCACGCUGAGUUUCACGCAGUUCUCCAUGGUGAGGCUAAGGACUUUGGCUAUUUU ...(((((..((((((..((((((..((((((((..((..((((((.((((.....))))...))))))..))))))))))......))))))..))))))..)))))...... ( -45.40) >DroGri_CAF1 17159 114 + 1 AACGUUGACCUCCUGUAGGUCACCCCAGCUGCGACCUAUUUUUAGCUUCACCCUGAGCGGCACACUUAAUUUGACACAGUUUUCCAUCGUCAGACUGAGGACUUUGGCCACUUU .....((((((.....))))))..((((((((...(((....)))..((.....)))))))...((((.((((((.............)))))).)))).....)))....... ( -24.72) >DroSec_CAF1 7523 114 + 1 CACGCUAAUCUCCUUUAAUUCACCCCAACUGCGUCCGAUCCUCAGCUUCACCUUGAGGGGCACGCUGAGUUUCACGCAGCUCUCCAUGGUGAUGCUAAGGACUUUGGCUAUUUU ...(((((..(((((....(((((....((((((..((..((((((....((.....))....))))))..))))))))........)))))....)))))..)))))...... ( -41.10) >DroSim_CAF1 7930 114 + 1 CACGCUAAUCUCCUUUAAUUCACCCCAACUGCGUCCGAUCCUCAGCUUCACCUUUAGGGGCACGCUGAGUUUCACGCAGCUCUCCAUGGUGAUGCUAAGAACUUUGGCUAUUUU ...(((((....(((....(((((....((((((..((..((((((....((.....))....))))))..))))))))........)))))....)))....)))))...... ( -34.50) >DroWil_CAF1 12288 114 + 1 UAUUCCUACUUCCUUCAAUGCACCCCAGCUGCGACCAAUCUUCAGUUUCACCUUGAGUGGUACACUCAGUUUGACGCAGCUUUCCAUGGUGAGACUUAAAACUUUGGCUAUUUU ....((..............((((..(((((((..............(((..(((((((...)))))))..))))))))))......))))..............))....... ( -22.42) >DroYak_CAF1 7678 114 + 1 CAUCCUAAACUCCUGUAAUUCACCCCAACUGCGUCCAAUCCUCAGCUUUACCUUGAGGGGCACGCUGAGUUUCACGCAGUUCUCCAUGGUGAGGCUAAGGACUUUGGCUAUCUU ....(((((.((((....((((((..((((((((...(..((((((....((.....))....))))))..).))))))))......))))))....)))).)))))....... ( -37.60) >consensus CACGCUAACCUCCUUUAAUUCACCCCAACUGCGUCCGAUCCUCAGCUUCACCUUGAGGGGCACGCUGAGUUUCACGCAGCUCUCCAUGGUGAGGCUAAGGACUUUGGCUAUUUU ...(((((..((((.....(((((..((((((((..((..((((((....((......))...))))))..))))))))))......))))).....))))..)))))...... (-24.53 = -26.37 + 1.84)


| Location | 8,531,395 – 8,531,509 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 82.92 |
| Mean single sequence MFE | -37.52 |
| Consensus MFE | -27.25 |
| Energy contribution | -27.50 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8531395 114 - 27905053 AAAAUAGCCAAAGUCCUUAGCCUCACCAUGGAGAACUGCGUGAAACUCAGCGUGCCACUCAAGGUGAAGCUGAGGAUCGGACGCAGUUGGGGUGAAUUUAAGGAAGUUAGCGUG ......((.((..(((((((..(((((......((((((((((..((((((..(((......)))...))))))..))..))))))))..)))))..)))))))..)).))... ( -45.60) >DroGri_CAF1 17159 114 - 1 AAAGUGGCCAAAGUCCUCAGUCUGACGAUGGAAAACUGUGUCAAAUUAAGUGUGCCGCUCAGGGUGAAGCUAAAAAUAGGUCGCAGCUGGGGUGACCUACAGGAGGUCAACGUU ....(((((..(((..(((.(((((.(.(((...(((...........)))...))))))))).))).))).....((((((((.......)))))))).....)))))..... ( -33.40) >DroSec_CAF1 7523 114 - 1 AAAAUAGCCAAAGUCCUUAGCAUCACCAUGGAGAGCUGCGUGAAACUCAGCGUGCCCCUCAAGGUGAAGCUGAGGAUCGGACGCAGUUGGGGUGAAUUAAAGGAGAUUAGCGUG ......((.((..(((((....(((((......((((((((((..((((((..(((......)))...))))))..))..))))))))..)))))....)))))..)).))... ( -40.00) >DroSim_CAF1 7930 114 - 1 AAAAUAGCCAAAGUUCUUAGCAUCACCAUGGAGAGCUGCGUGAAACUCAGCGUGCCCCUAAAGGUGAAGCUGAGGAUCGGACGCAGUUGGGGUGAAUUAAAGGAGAUUAGCGUG ......((.((..(((((....(((((......((((((((((..((((((..(((......)))...))))))..))..))))))))..)))))....)))))..)).))... ( -37.10) >DroWil_CAF1 12288 114 - 1 AAAAUAGCCAAAGUUUUAAGUCUCACCAUGGAAAGCUGCGUCAAACUGAGUGUACCACUCAAGGUGAAACUGAAGAUUGGUCGCAGCUGGGGUGCAUUGAAGGAAGUAGGAAUA .............(((((..(((((((......(((((((((((.((.(((.((((......))))..)))..)).)))).)))))))..)))).......)))..)))))... ( -29.21) >DroYak_CAF1 7678 114 - 1 AAGAUAGCCAAAGUCCUUAGCCUCACCAUGGAGAACUGCGUGAAACUCAGCGUGCCCCUCAAGGUAAAGCUGAGGAUUGGACGCAGUUGGGGUGAAUUACAGGAGUUUAGGAUG .......(((((.((((.....(((((......((((((((((..((((((.((((......))))..))))))..))..))))))))..))))).....)))).))).))... ( -39.80) >consensus AAAAUAGCCAAAGUCCUUAGCCUCACCAUGGAGAACUGCGUGAAACUCAGCGUGCCCCUCAAGGUGAAGCUGAGGAUCGGACGCAGUUGGGGUGAAUUAAAGGAGGUUAGCGUG .............((((.....(((((......((((((((((..((((((..(((......)))...))))))..))..))))))))..))))).....)))).......... (-27.25 = -27.50 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:40 2006