
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,495,901 – 8,496,061 |
| Length | 160 |
| Max. P | 0.954491 |

| Location | 8,495,901 – 8,496,021 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.68 |
| Mean single sequence MFE | -37.43 |
| Consensus MFE | -26.61 |
| Energy contribution | -26.70 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.23 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
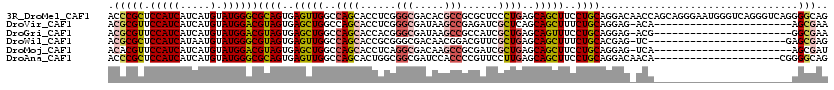
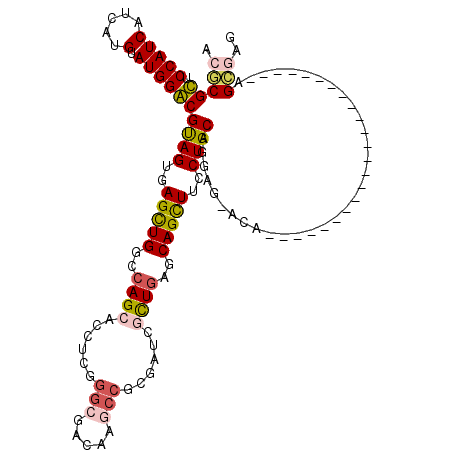
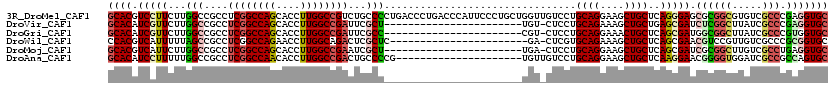
>3R_DroMel_CAF1 8495901 120 + 27905053 ACCCGCUCCAUCAUCAUGUAUGGGCGCAGUGAGUUGGCCAGCACCUCGGGCGACACGCCGCGCUCCCUGAGCAGCUUCCUGCAGGACAACCAGCAGGGAAUGGGUCAGGGUCAGGGGCAG ....(((((.............(((((.(((.((((.((........)).)))).))).)))))((((((.(...(((((((.((....)).)))))))...).))))))...))))).. ( -54.90) >DroVir_CAF1 14501 96 + 1 ACGCGUUCCAUCAUCAUGUAUGGACGUAGUGAGCUGGCCAGCACCUCGGGCGAUAAGCCGAGAUCGCUCAGCAGCUUUCUGCAGGAG-ACA-----------------------AGCGAA .(((((.(((((.....).))))))((((.((((((...(((..(((((.(.....))))))...)))...)))))).)))).(...-.).-----------------------.))).. ( -34.50) >DroGri_CAF1 15479 96 + 1 ACGCGUUCCAUCAUCAUGUAUGGACGUAGUGAGCUGGCCAGCACCACGGGCGAUAAGCCGCCAUCGCUGAGCAGUUUCCUGCAGGAG-ACG-----------------------GGCGAA .(((((.(((((.....).))))))((((..(((((..((((......((((......))))...))))..)))))..)))).(...-.).-----------------------.))).. ( -34.90) >DroWil_CAF1 4343 96 + 1 ACGCGCUCCAUCAUAAUGUAUGGGCGUAGUGAGUUGGCCAGCACCGCGGGCGACAACGGACGUUCGCUGAGCAGCUUUCUGCACGAG-UC-----------------------GAGCGAG .(((((.(((((.....).))))))((((.((((((..((((...(((..((....))..)))..))))..)))))).)))).....-..-----------------------..))).. ( -33.50) >DroMoj_CAF1 12474 96 + 1 ACACGUUCCAUCAUCAUGUAUGGACGUAGUGAGCUGGCCAGCACCUCAGGCGACAAGCCGCGAUCGCUGAGCAGCUUCCUGCAGGAG-UCA-----------------------AGCGAU ...(((((((((.....).))))).((((..(((((..((((...((.(((.....)))..))..))))..)))))..)))).....-...-----------------------.))).. ( -32.80) >DroAna_CAF1 4434 99 + 1 ACCCGCUCCAUCAUCAUGUAUGGGCGCAGUGAGUUGGCCAGCACUGGCGGCGAUCCACCCCGUUCCUUGAGCAGCUUCCUGCAGGACAACA---------------------CGGGGCAG .(((((.(((((.....).))))))((((..(((((((((....))))((((........)))).......)))))..)))).........---------------------.))).... ( -34.00) >consensus ACGCGCUCCAUCAUCAUGUAUGGACGUAGUGAGCUGGCCAGCACCUCGGGCGACAAGCCGCGAUCGCUGAGCAGCUUCCUGCAGGAG_ACA_______________________AGCGAG .(((((.(((((.....).))))))((((..(((((..((((......(((.....)))......))))..)))))..)))).................................))).. (-26.61 = -26.70 + 0.09)
| Location | 8,495,941 – 8,496,061 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.17 |
| Mean single sequence MFE | -44.87 |
| Consensus MFE | -27.43 |
| Energy contribution | -26.85 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.920641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
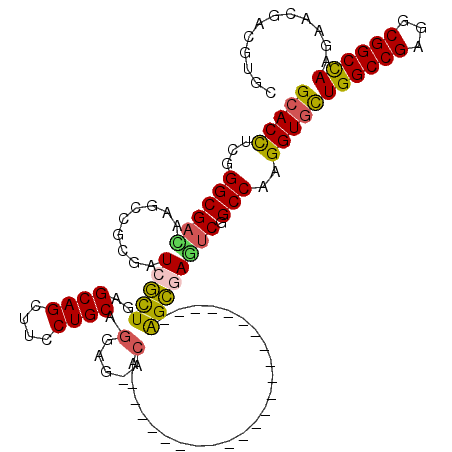
>3R_DroMel_CAF1 8495941 120 + 27905053 GCACCUCGGGCGACACGCCGCGCUCCCUGAGCAGCUUCCUGCAGGACAACCAGCAGGGAAUGGGUCAGGGUCAGGGGCAGACGGCCAAGGUGCUGGCCGAGGCGGCCAAGAAGGACGUGC ((((((.((.((....(((.(...((((((.(...(((((((.((....)).)))))))...).))))))...).)))...)).)).))))))((((((...))))))............ ( -59.70) >DroVir_CAF1 14541 96 + 1 GCACCUCGGGCGAUAAGCCGAGAUCGCUCAGCAGCUUUCUGCAGGAG-ACA-----------------------AGCGAAUCGGCCAAGGUGCUGGCCGAGGCGGCCAAGAACGAUGUGC ((((.(((((((....(((((..(((((..((((....)))).(...-.).-----------------------))))).))))).....))))(((((...))))).....))).)))) ( -42.70) >DroGri_CAF1 15519 96 + 1 GCACCACGGGCGAUAAGCCGCCAUCGCUGAGCAGUUUCCUGCAGGAG-ACG-----------------------GGCGAAUCGGCCAAGGUGCUGGCCGAGGCGGCCAAGAACGAUGUGC ((((..((..(.....((((((.(((((..((((....)))).(...-.).-----------------------))))).(((((((......)))))))))))))...)..))..)))) ( -47.00) >DroWil_CAF1 4383 96 + 1 GCACCGCGGGCGACAACGGACGUUCGCUGAGCAGCUUUCUGCACGAG-UC-----------------------GAGCGAGUCUGCCAAGGUUCUGGCCGAGGCGGCUAAAAAUGACGUGG ...(((((............(((((((((.((((....)))).).))-.)-----------------------)))))((.(((((..(((....)))..)))))))........))))) ( -34.30) >DroMoj_CAF1 12514 96 + 1 GCACCUCAGGCGACAAGCCGCGAUCGCUGAGCAGCUUCCUGCAGGAG-UCA-----------------------AGCGAUUCGGCCAAGGUGCUGGCCGAGGCGGCCAAGAAUGACGUGC ((((.(((..(.....(((((..(((((((((((....)))).....-)).-----------------------)))))((((((((......)))))))))))))...)..))).)))) ( -41.80) >DroAna_CAF1 4474 99 + 1 GCACUGGCGGCGAUCCACCCCGUUCCUUGAGCAGCUUCCUGCAGGACAACA---------------------CGGGGCAGUCGGCCAAGGUGUUGGCCGAGGCGGCCAAAAAGGAUGUGC ((((((((.((......((((((((((...((((....))))))))....)---------------------)))))...((((((((....)))))))).)).))))........)))) ( -43.70) >consensus GCACCUCGGGCGACAAGCCGCGAUCGCUGAGCAGCUUCCUGCAGGAG_ACA_______________________AGCGAGUCGGCCAAGGUGCUGGCCGAGGCGGCCAAGAACGACGUGC (((((...((((((.........(((((..((((....)))).(.....)........................)))))))).)))..)))))((((((...))))))............ (-27.43 = -26.85 + -0.58)

| Location | 8,495,941 – 8,496,061 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.17 |
| Mean single sequence MFE | -43.57 |
| Consensus MFE | -26.71 |
| Energy contribution | -27.43 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
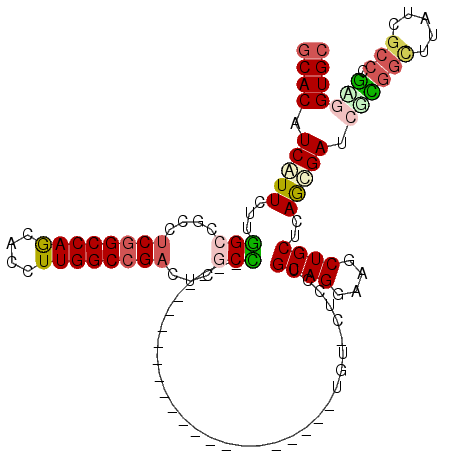
>3R_DroMel_CAF1 8495941 120 - 27905053 GCACGUCCUUCUUGGCCGCCUCGGCCAGCACCUUGGCCGUCUGCCCCUGACCCUGACCCAUUCCCUGCUGGUUGUCCUGCAGGAAGCUGCUCAGGGAGCGCGGCGUGUCGCCCGAGGUGC ............((((((...))))))(((((((((.((.((((((((..((((((.(..((.(((((.((....)).))))).))..).)))))))).).)))).).)).))))))))) ( -55.00) >DroVir_CAF1 14541 96 - 1 GCACAUCGUUCUUGGCCGCCUCGGCCAGCACCUUGGCCGAUUCGCU-----------------------UGU-CUCCUGCAGAAAGCUGCUGAGCGAUCUCGGCUUAUCGCCCGAGGUGC ............((((((...))))))(((((((((.((((..(((-----------------------((.-(((..((((....)))).))))).....)))..)))).))))))))) ( -40.90) >DroGri_CAF1 15519 96 - 1 GCACAUCGUUCUUGGCCGCCUCGGCCAGCACCUUGGCCGAUUCGCC-----------------------CGU-CUCCUGCAGGAAACUGCUCAGCGAUGGCGGCUUAUCGCCCGUGGUGC ............((((((...))))))(((((.(((.((((..(((-----------------------.((-(((((((((....))))..)).)).))))))..)))).))).))))) ( -44.10) >DroWil_CAF1 4383 96 - 1 CCACGUCAUUUUUAGCCGCCUCGGCCAGAACCUUGGCAGACUCGCUC-----------------------GA-CUCGUGCAGAAAGCUGCUCAGCGAACGUCCGUUGUCGCCCGCGGUGC ..............(((((....(((((....))))).(((.((..(-----------------------(.-.((((((((....))))...)))).))..))..)))....))))).. ( -28.40) >DroMoj_CAF1 12514 96 - 1 GCACGUCAUUCUUGGCCGCCUCGGCCAGCACCUUGGCCGAAUCGCU-----------------------UGA-CUCCUGCAGGAAGCUGCUCAGCGAUCGCGGCUUGUCGCCUGAGGUGC ((((.(((.....((((((.((((((((....))))))))((((((-----------------------.((-.....((((....)))))))))))).)))))).......))).)))) ( -44.80) >DroAna_CAF1 4474 99 - 1 GCACAUCCUUUUUGGCCGCCUCGGCCAACACCUUGGCCGACUGCCCCG---------------------UGUUGUCCUGCAGGAAGCUGCUCAAGGAACGGGGUGGAUCGCCGCCAGUGC ((((........((((.((.((((((((....))))))))(..(((((---------------------(....((((((((....))))...))))))))))..)...)).)))))))) ( -48.20) >consensus GCACAUCAUUCUUGGCCGCCUCGGCCAGCACCUUGGCCGACUCGCC_______________________UGU_CUCCUGCAGGAAGCUGCUCAGCGAUCGCGGCUUAUCGCCCGAGGUGC ((((.(((((...(((....((((((((....))))))))...)))................................((((....))))..))))).((((((.....))).))))))) (-26.71 = -27.43 + 0.73)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:35 2006