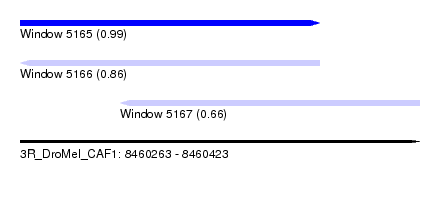
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,460,263 – 8,460,423 |
| Length | 160 |
| Max. P | 0.986990 |

| Location | 8,460,263 – 8,460,383 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.77 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -31.97 |
| Energy contribution | -32.30 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
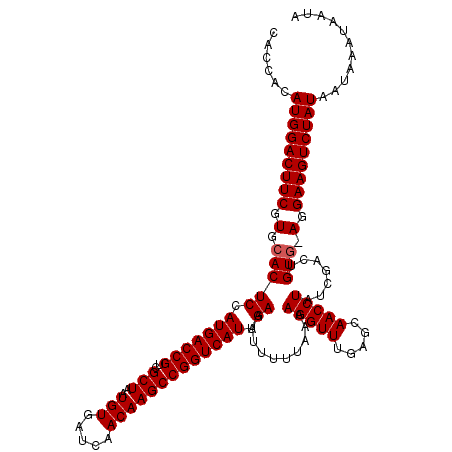
>3R_DroMel_CAF1 8460263 120 + 27905053 CACCACAUGGACUUCGUGCACUCUAUGACCGUCGCUAAUGUGAUUAACAAGCCGGUCAUUGAAAUUUUUAAAAGGUUUGAGCAACCUAUCGACUGUAAAGGAAGUCUAUAAUAAAUAAUA ......(((((((((.((((.((.(((((((..(((..(((.....))))))))))))).............(((((.....)))))...)).))))...)))))))))........... ( -31.40) >DroSec_CAF1 6166 119 + 1 CACCACAUGGACUUCGUGCACUCCAUGACCGUCGCUAAUGUGAUCAACAAGCCGGUCAUUGAAAUUUUUAAAAGGUUUGAGCAACCUAUCGACUGUG-AGGAAGUCUAUAAUAAAUAAUA ......(((((((((.(.(((((.(((((((..(((..(((.....))))))))))))).))..........(((((.....))))).......)))-).)))))))))........... ( -32.90) >DroSim_CAF1 6224 119 + 1 CACCACAUGGACUUCGUGCACUCCAUGACCGUCGCUAAUGUGAUCAACAAGCCGGUCAUUGAAAUUUUUAAAAGGUUUGAGCAACCUAUCGACUGUG-AGGAAGUCUAUAAUAAAUAAUA ......(((((((((.(.(((((.(((((((..(((..(((.....))))))))))))).))..........(((((.....))))).......)))-).)))))))))........... ( -32.90) >consensus CACCACAUGGACUUCGUGCACUCCAUGACCGUCGCUAAUGUGAUCAACAAGCCGGUCAUUGAAAUUUUUAAAAGGUUUGAGCAACCUAUCGACUGUG_AGGAAGUCUAUAAUAAAUAAUA ......(((((((((.(.(((((.(((((((..(((..(((.....))))))))))))).))..........(((((.....))))).......))).).)))))))))........... (-31.97 = -32.30 + 0.33)

| Location | 8,460,263 – 8,460,383 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.77 |
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -31.71 |
| Energy contribution | -31.27 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.73 |
| Structure conservation index | 1.02 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8460263 120 - 27905053 UAUUAUUUAUUAUAGACUUCCUUUACAGUCGAUAGGUUGCUCAAACCUUUUAAAAAUUUCAAUGACCGGCUUGUUAAUCACAUUAGCGACGGUCAUAGAGUGCACGAAGUCCAUGUGGUG .......(((((((((((((...(((....((.(((((.....))))).)).......((.(((((((..((((((((...))))))))))))))).)))))...))))))..))))))) ( -30.00) >DroSec_CAF1 6166 119 - 1 UAUUAUUUAUUAUAGACUUCCU-CACAGUCGAUAGGUUGCUCAAACCUUUUAAAAAUUUCAAUGACCGGCUUGUUGAUCACAUUAGCGACGGUCAUGGAGUGCACGAAGUCCAUGUGGUG .......(((((((((((((..-(((....((.(((((.....))))).)).......((.(((((((..((((((((...))))))))))))))).)))))...))))))..))))))) ( -31.60) >DroSim_CAF1 6224 119 - 1 UAUUAUUUAUUAUAGACUUCCU-CACAGUCGAUAGGUUGCUCAAACCUUUUAAAAAUUUCAAUGACCGGCUUGUUGAUCACAUUAGCGACGGUCAUGGAGUGCACGAAGUCCAUGUGGUG .......(((((((((((((..-(((....((.(((((.....))))).)).......((.(((((((..((((((((...))))))))))))))).)))))...))))))..))))))) ( -31.60) >consensus UAUUAUUUAUUAUAGACUUCCU_CACAGUCGAUAGGUUGCUCAAACCUUUUAAAAAUUUCAAUGACCGGCUUGUUGAUCACAUUAGCGACGGUCAUGGAGUGCACGAAGUCCAUGUGGUG .......(((((((((((((...(((....((.(((((.....))))).)).......((.(((((((..((((((((...))))))))))))))).)))))...))))))..))))))) (-31.71 = -31.27 + -0.44)

| Location | 8,460,303 – 8,460,423 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.10 |
| Mean single sequence MFE | -29.73 |
| Consensus MFE | -28.80 |
| Energy contribution | -29.13 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
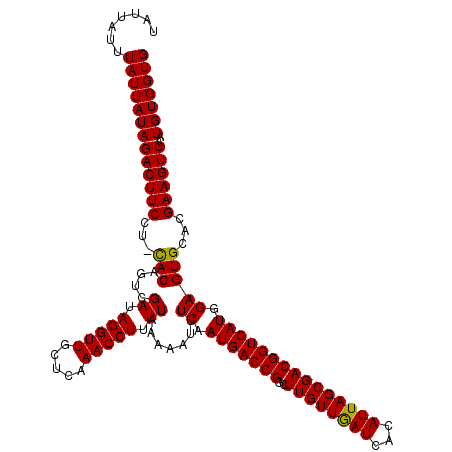
>3R_DroMel_CAF1 8460303 120 - 27905053 AAAGACCGGACGUAGUCGAUAAUGGCGGGCUGGUACUCGAUAUUAUUUAUUAUAGACUUCCUUUACAGUCGAUAGGUUGCUCAAACCUUUUAAAAAUUUCAAUGACCGGCUUGUUAAUCA ...(((........)))(((..((((((((((((....((.(((..........((((........))))((.(((((.....))))).))...))).))....))))))))))))))). ( -28.60) >DroSec_CAF1 6206 119 - 1 AAAGACCAGACGUUGUCGAUCAUGGCGGGCUGGUACUCCAUAUUAUUUAUUAUAGACUUCCU-CACAGUCGAUAGGUUGCUCAAACCUUUUAAAAAUUUCAAUGACCGGCUUGUUGAUCA ...(((........)))(((((..((((((((((....................((((....-...))))((.(((((.....))))).)).............))))))))))))))). ( -30.20) >DroSim_CAF1 6264 119 - 1 AAAGACCAGACGUAGUCGAUCAUGGCGGGCUGGUACUCCAUAUUAUUUAUUAUAGACUUCCU-CACAGUCGAUAGGUUGCUCAAACCUUUUAAAAAUUUCAAUGACCGGCUUGUUGAUCA ...(((........)))(((((..((((((((((....................((((....-...))))((.(((((.....))))).)).............))))))))))))))). ( -30.40) >consensus AAAGACCAGACGUAGUCGAUCAUGGCGGGCUGGUACUCCAUAUUAUUUAUUAUAGACUUCCU_CACAGUCGAUAGGUUGCUCAAACCUUUUAAAAAUUUCAAUGACCGGCUUGUUGAUCA ...(((........)))(((((..((((((((((....................((((........))))((.(((((.....))))).)).............))))))))))))))). (-28.80 = -29.13 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:16 2006