
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,459,943 – 8,460,143 |
| Length | 200 |
| Max. P | 0.988772 |

| Location | 8,459,943 – 8,460,063 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.08 |
| Mean single sequence MFE | -38.54 |
| Consensus MFE | -34.60 |
| Energy contribution | -35.20 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
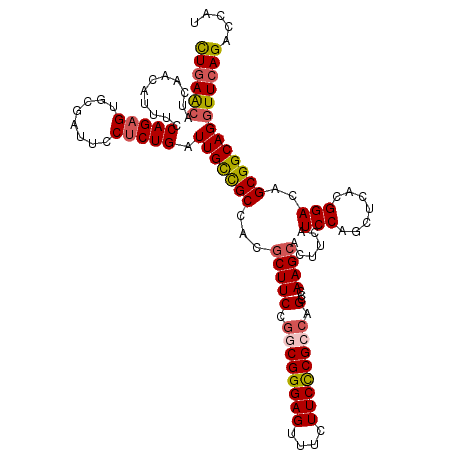
>3R_DroMel_CAF1 8459943 120 + 27905053 CUGAACAUUAACAUUUCCAGAGUGCGAUUCCUCUGAUUGCCGCCACGCUUCCAGCGGGAGUUUCUUCCCGCCAGCCCAAGCACUUCAUCCAGCUCACGGACAGCGGCAGGUUCAGACCAU ((((((...........(((((........))))).(((((((...((((...(((((((....)))))))......))))......(((.......)))..)))))))))))))..... ( -41.10) >DroSec_CAF1 5846 120 + 1 CUGAACAUCAACACUUCCAGAGUGCGAUUCCUCUGAUUGCCGCCACGCUUCCGGCGGGAGUUUCUUCCCGGCAGCCCAAGCACUUCAUCCAGCUGACGGACAGCGGCAGGUUCAGACCAU ((((((...........(((((........))))).(((((((...(((.((((..((.....))..)))).)))((.(((..........)))...))...)))))))))))))..... ( -41.50) >DroSim_CAF1 5904 120 + 1 UUGAACAUCAACACUUUCAGAGUGCGAUUCCUCUGAUUGCCGCCACGCUUCCGGCGGGAGUUUCUUCCCGGCAGCCCAAGCACUUCAUCCAGCUGACGGACAGCGGCAGGUUCAGACCAU ((((((..........((((((........))))))(((((((...(((.((((..((.....))..)))).)))((.(((..........)))...))...)))))))))))))..... ( -40.50) >DroEre_CAF1 6001 120 + 1 CUAAGCAUCAACAUUUCCAGAGUGCUAUUCCUCUGAUUGCUGCCACCCUUCCGGCGGGAGUUUCUUCCCGCCAGCCAAAGCACUUCAUCCAGCUCACGGACAGCGGCAGGUUCAGCCUGU .................(((((........)))))..(((((((........((((((((....)))))))).((....))................)).)))))(((((.....))))) ( -37.90) >DroYak_CAF1 5962 120 + 1 CUGAACAUUAACAUUUCCAGAGUGCAAUUCCACUGAUUGUCGCCACGCUUCCGGCGGGAGUUUCUUCUCGCCAGCCAAAGCACUUCAUCCAGCUCUCGGACAGCGGCAGAUUCAGCUUGU (((((...............((((......))))..(((((((...(((((.((((((((....)))))))).)...))))......(((.......)))..))))))).)))))..... ( -31.70) >consensus CUGAACAUCAACAUUUCCAGAGUGCGAUUCCUCUGAUUGCCGCCACGCUUCCGGCGGGAGUUUCUUCCCGCCAGCCCAAGCACUUCAUCCAGCUCACGGACAGCGGCAGGUUCAGACCAU ((((((...........(((((........))))).(((((((...(((((.((((((((....)))))))).)...))))......(((.......)))..)))))))))))))..... (-34.60 = -35.20 + 0.60)


| Location | 8,459,943 – 8,460,063 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.08 |
| Mean single sequence MFE | -45.12 |
| Consensus MFE | -40.32 |
| Energy contribution | -41.12 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8459943 120 - 27905053 AUGGUCUGAACCUGCCGCUGUCCGUGAGCUGGAUGAAGUGCUUGGGCUGGCGGGAAGAAACUCCCGCUGGAAGCGUGGCGGCAAUCAGAGGAAUCGCACUCUGGAAAUGUUAAUGUUCAG .....((((((.((((((((((((.....))))).....((((...(..((((((......))))))..)))))..))))))).((((((........))))))..........)))))) ( -48.60) >DroSec_CAF1 5846 120 - 1 AUGGUCUGAACCUGCCGCUGUCCGUCAGCUGGAUGAAGUGCUUGGGCUGCCGGGAAGAAACUCCCGCCGGAAGCGUGGCGGCAAUCAGAGGAAUCGCACUCUGGAAGUGUUGAUGUUCAG .....(((((((((((((..((((.(((((.((........)).))))).(((((......))))).))))...)))))))...((((((........))))))..........)))))) ( -44.70) >DroSim_CAF1 5904 120 - 1 AUGGUCUGAACCUGCCGCUGUCCGUCAGCUGGAUGAAGUGCUUGGGCUGCCGGGAAGAAACUCCCGCCGGAAGCGUGGCGGCAAUCAGAGGAAUCGCACUCUGAAAGUGUUGAUGUUCAA ......((((((((((((..((((.(((((.((........)).))))).(((((......))))).))))...)))))))...((((((........))))))..........))))). ( -43.80) >DroEre_CAF1 6001 120 - 1 ACAGGCUGAACCUGCCGCUGUCCGUGAGCUGGAUGAAGUGCUUUGGCUGGCGGGAAGAAACUCCCGCCGGAAGGGUGGCAGCAAUCAGAGGAAUAGCACUCUGGAAAUGUUGAUGCUUAG ..(((((.(((((((((((.(((...(((..((........))..)))(((((((......))))))))))..))))))))...((((((........))))))....))).).)))).. ( -51.10) >DroYak_CAF1 5962 120 - 1 ACAAGCUGAAUCUGCCGCUGUCCGAGAGCUGGAUGAAGUGCUUUGGCUGGCGAGAAGAAACUCCCGCCGGAAGCGUGGCGACAAUCAGUGGAAUUGCACUCUGGAAAUGUUAAUGUUCAG .....((((((..(((((.(((((.....))))).....((((((((.((.(((......)))))))).))))))))))((((.((((.(........).))))...))))...)))))) ( -37.40) >consensus AUGGUCUGAACCUGCCGCUGUCCGUGAGCUGGAUGAAGUGCUUGGGCUGGCGGGAAGAAACUCCCGCCGGAAGCGUGGCGGCAAUCAGAGGAAUCGCACUCUGGAAAUGUUGAUGUUCAG .....(((((((((((((.(((((.....))))).....((((...(((((((((......))))))))))))))))))))...((((((........))))))..........)))))) (-40.32 = -41.12 + 0.80)


| Location | 8,460,023 – 8,460,143 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.00 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -24.89 |
| Energy contribution | -25.53 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8460023 120 + 27905053 CACUUCAUCCAGCUCACGGACAGCGGCAGGUUCAGACCAUUCGAUUUCGGGAUCCUGAGGAACUUGAUCAACUACAGGAGCCUCACACCUCCCGACUACCCGCUACAUAACGUCCGCCCC ................((((((((((..(((....)))........((((((...(((((..((((........))))..)))))....))))))....))))).......))))).... ( -35.71) >DroSec_CAF1 5926 120 + 1 CACUUCAUCCAGCUGACGGACAGCGGCAGGUUCAGACCAUUCGAUUUCGGGAUCCUGAGGAACUUGAUCAACUACAGGAGCCUCACACCUCCCGACUACCCGCUGCAUAACGUCCGCCCU ...........((.((((..((((((..(((....)))........((((((...(((((..((((........))))..)))))....))))))....)))))).....)))).))... ( -40.10) >DroSim_CAF1 5984 120 + 1 CACUUCAUCCAGCUGACGGACAGCGGCAGGUUCAGACCAUUCGAUUUCGGGAUCCUAAAGAACUUGAUCAACUACAGGAGCCUCACACCUCCCGACUACCCGCUGCAUAACGUCCGCCCU ...........((.((((..(((((((((((((.((....))((((....)))).....)))))))........(.((((........)))).).....)))))).....)))).))... ( -30.60) >DroEre_CAF1 6081 120 + 1 CACUUCAUCCAGCUCACGGACAGCGGCAGGUUCAGCCUGUUCGAUUUCGGUAUCCUAAAAAACUUGAUCUACUACAGGAGUCUUACACCCCCCGAUUACCCGCUACAUAAUGUCCACCCC .................(((((((((((((.....))))...((((((.(((....................))).)))))).................)))).......)))))..... ( -21.86) >DroYak_CAF1 6042 120 + 1 CACUUCAUCCAGCUCUCGGACAGCGGCAGAUUCAGCUUGUUCGAUUUCGGGAUCCUGAGAAACUUGAUCUACUACAGGAGGCUAACACCUCCCGAUUACCCGCUACAUAACGUCCGCCCC ................((((((((((.((((.(((...(((...((((((....))))))))))))))))....(.(((((......))))).).....))))).......))))).... ( -28.71) >consensus CACUUCAUCCAGCUCACGGACAGCGGCAGGUUCAGACCAUUCGAUUUCGGGAUCCUGAGGAACUUGAUCAACUACAGGAGCCUCACACCUCCCGACUACCCGCUACAUAACGUCCGCCCC ................((((((((((..((......))........((((((...(((((..((((........))))..)))))....))))))....))))).......))))).... (-24.89 = -25.53 + 0.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:13 2006