
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,450,491 – 8,450,585 |
| Length | 94 |
| Max. P | 0.935765 |

| Location | 8,450,491 – 8,450,585 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.09 |
| Mean single sequence MFE | -34.33 |
| Consensus MFE | -31.40 |
| Energy contribution | -31.10 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
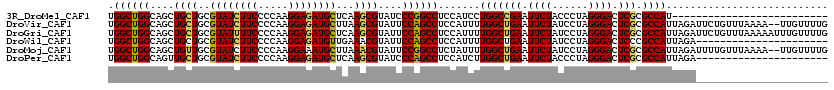
>3R_DroMel_CAF1 8450491 94 + 27905053 --------------------------AUGGCGCGAGUCCCUAGGGUAGAAUUCGGCCAGGAUGGAGGCCGGGAUACGCUUGAGCAUCUCCUUGGGGAAGAUACGCAGCAGCUGCCAGCCA --------------------------.((((((((.(((((((((.(((.(((((((........)))))))....((....)).))))))))))))...).))).((....))..)))) ( -40.60) >DroVir_CAF1 3248 118 + 1 CAAAACAA--UUUUAAACAGAAUCUAAUGGCGCGAGUCCCUAGGAUAGAAUUCAGCCAAAAUGGAGGCUGGAAUACGCUUAAGCAUCUCCUUGGGGAAGAUACGCAGCAGCUGCCAGCCA ........--.................((((((((.((((((((..(((.(((((((........)))))))....((....)).))).))))))))...).))).((....))..)))) ( -35.00) >DroGri_CAF1 2289 120 + 1 CAAAACAAAUUUUUAAACAGAAUCUAAUGGCGCGAGUCCCUAGGAUAGAAUUCAGCCAAAAUGGAGGCUGGAAUACGCUUGAGCAUCUCCUUGGGGAAAAUACGCAGCAGCUGCCAGCCA ...........................((((((((.((((((((..(((.(((((((........)))))))....((....)).))).))))))))...).))).((....))..)))) ( -35.00) >DroWil_CAF1 1 98 + 1 ----------------------UCUAAUGGCGGGAGUCCCUAGGAUAGAAUUCAGCCAAAAUGGAGGCUGGAAUACGUUUCAACAUCUCCUUGGGGAAGAUACGCAGCAGCUGCCAGCCA ----------------------.....((((.((..((((((((..(((.(((((((........)))))))....((....)).))).))))))))......((....))..)).)))) ( -30.90) >DroMoj_CAF1 2399 118 + 1 CAAAACAA--UUUUAAACAAAAUCUAAUGGCGCGAGUCCCUAGGAUAGAAUUCAGCCAAAAUAGAGGCCGGAAUACGUUUAAGCAUUUCCUUGGGGAAGAUACGCAACAGCUGCCAGCCA ........--.................((((.....(((((((((..(.((((.(((........)))..)))).)((....))...))).))))))......(((.....)))..)))) ( -25.40) >DroPer_CAF1 1 98 + 1 ----------------------UCUAAUGGCGCGAGUCCCUAGGGUAGAAUUCAGCCAAGAUGGAGGCUGGGAUACGCUUGAGCAUCUCCUUGGGGAAGAUACGCAGCAACUGCCAGCCA ----------------------.....((((((((.(((((((((.(((.(((((((........)))))))....((....)).))))))))))))...).))).((....))..)))) ( -39.10) >consensus ______________________UCUAAUGGCGCGAGUCCCUAGGAUAGAAUUCAGCCAAAAUGGAGGCUGGAAUACGCUUGAGCAUCUCCUUGGGGAAGAUACGCAGCAGCUGCCAGCCA ...........................((((((((.((((((((......(((((((........)))))))....((....)).....))))))))...).))).((....))..)))) (-31.40 = -31.10 + -0.30)


| Location | 8,450,491 – 8,450,585 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.09 |
| Mean single sequence MFE | -33.27 |
| Consensus MFE | -30.90 |
| Energy contribution | -30.60 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
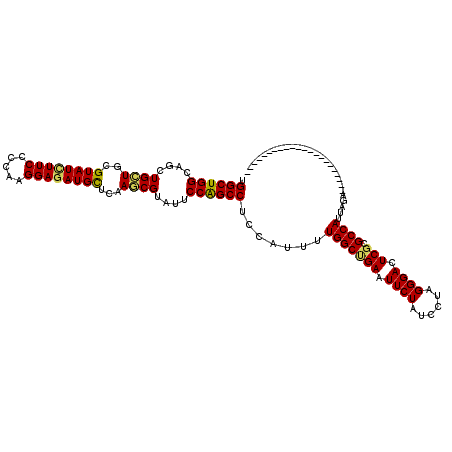
>3R_DroMel_CAF1 8450491 94 - 27905053 UGGCUGGCAGCUGCUGCGUAUCUUCCCCAAGGAGAUGCUCAAGCGUAUCCCGGCCUCCAUCCUGGCCGAAUUCUACCCUAGGGACUCGCGCCAU-------------------------- ((((..((((...))))((((((((.....))))))))....(((..((((((((........))))(....).......))))..))))))).-------------------------- ( -37.30) >DroVir_CAF1 3248 118 - 1 UGGCUGGCAGCUGCUGCGUAUCUUCCCCAAGGAGAUGCUUAAGCGUAUUCCAGCCUCCAUUUUGGCUGAAUUCUAUCCUAGGGACUCGCGCCAUUAGAUUCUGUUUAAAA--UUGUUUUG .((((((....((((..((((((((.....))))))))...))))....)))))).......(((((((.((((......)))).))).)))).................--........ ( -34.20) >DroGri_CAF1 2289 120 - 1 UGGCUGGCAGCUGCUGCGUAUUUUCCCCAAGGAGAUGCUCAAGCGUAUUCCAGCCUCCAUUUUGGCUGAAUUCUAUCCUAGGGACUCGCGCCAUUAGAUUCUGUUUAAAAAUUUGUUUUG ((((.(((....)))(((.....((((..((((.((((....))))....(((((........))))).......)))).))))..)))))))........................... ( -33.20) >DroWil_CAF1 1 98 - 1 UGGCUGGCAGCUGCUGCGUAUCUUCCCCAAGGAGAUGUUGAAACGUAUUCCAGCCUCCAUUUUGGCUGAAUUCUAUCCUAGGGACUCCCGCCAUUAGA---------------------- ((((.((..((....))........(((.((((.((((....))))....(((((........))))).......)))).)))...)).)))).....---------------------- ( -29.10) >DroMoj_CAF1 2399 118 - 1 UGGCUGGCAGCUGUUGCGUAUCUUCCCCAAGGAAAUGCUUAAACGUAUUCCGGCCUCUAUUUUGGCUGAAUUCUAUCCUAGGGACUCGCGCCAUUAGAUUUUGUUUAAAA--UUGUUUUG .((((((....((((..((((.((((....))))))))...))))....))))))((((...(((((((.((((......)))).))).)))).))))............--........ ( -30.50) >DroPer_CAF1 1 98 - 1 UGGCUGGCAGUUGCUGCGUAUCUUCCCCAAGGAGAUGCUCAAGCGUAUCCCAGCCUCCAUCUUGGCUGAAUUCUACCCUAGGGACUCGCGCCAUUAGA---------------------- .((((((....(((((.((((((((.....)))))))).).))))....))))))....((((((((((.((((......)))).))).))))..)))---------------------- ( -35.30) >consensus UGGCUGGCAGCUGCUGCGUAUCUUCCCCAAGGAGAUGCUCAAGCGUAUUCCAGCCUCCAUUUUGGCUGAAUUCUAUCCUAGGGACUCGCGCCAUUAGA______________________ .((((((....((((..((((((((.....))))))))...))))....)))))).......(((((((.((((......)))).))).))))........................... (-30.90 = -30.60 + -0.30)

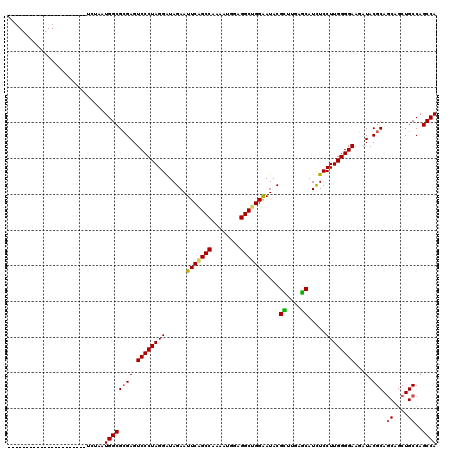
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:02 2006