| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,400,702 – 8,400,803 |
| Length | 101 |
| Max. P | 0.984373 |

| Location | 8,400,702 – 8,400,803 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.99 |
| Mean single sequence MFE | -39.43 |
| Consensus MFE | -23.14 |
| Energy contribution | -26.42 |
| Covariance contribution | 3.28 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
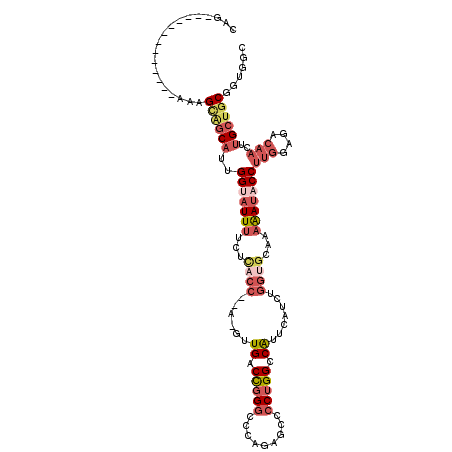
>3R_DroMel_CAF1 8400702 101 + 27905053 GCCAACGCAGCAAGUUGUCUCCAAGGUAUUUUUGCACCAGAUGAAUGACCAGGGGCUCUGCGCCCGGUCAAC-U--GGUGAGAAAAUUCCAAUGGCACUUU-------------UUG ((((..((((....))))......((.(((((..((((((.....(((((..((((.....))))))))).)-)--))))..))))).))..)))).....-------------... ( -35.80) >DroSec_CAF1 62553 102 + 1 GCCAACGCAGCAAGUUGUCUCCAAGGUAUUUUUGCACCAGAUGAAUGGCCAGGAGCUUUGGGCCCGGUCAGGAA--GGAAAGCAAAUACCAAUGCUGCUUU-------------CUG ......((((((..(((....)))((((((..(((.((.......(((((.((.((.....)))))))))....--))...)))))))))..))))))...-------------... ( -29.20) >DroSim_CAF1 61720 101 + 1 GCCAACGCAGCAAGUUGUCUCCAAGGUAUUUUUGCACCAGAUGAAUGGCCAGGGGCUCUGGGCCCGGUCAAC-U--GGUGAGGAAAUACCAAUGCUGCUUU-------------CUG ......((((((..(((....)))((((((((..((((((.....(((((.(((.(....).)))))))).)-)--))))..))))))))..))))))...-------------... ( -42.70) >DroEre_CAF1 64520 101 + 1 GCCACCGCAGCAAGUUGUCUCCAAGGUAUUUUUGCACCAGAUGAAUGGCCAGGUCCUCUGGGCCCGGUCAAC-U--GGUGAGAAAAUACCAAUGCUGCUUU-------------CUG ......((((((..(((....)))((((((((..((((((.....(((((.(((((...))))).))))).)-)--))))..))))))))..))))))...-------------... ( -43.30) >DroYak_CAF1 64347 103 + 1 GCCACCGCAGCAAGUUGUCUCCAAGGUAUUUUUGCACCAGAUGAAUGGCCAGGGGCUCUGGGCCCGGUCAAC-CUGGGUGGGAAAAUACCAAUGCUGCUUU-------------CUG ......((((((..(((....)))(((((((((.((((....(..(((((.(((.(....).))))))))..-)..)))).)))))))))..))))))...-------------... ( -41.80) >DroPer_CAF1 65175 107 + 1 ACUGCCGCCGCAAUGUGUCUCUGAGGUGUCUUUGGGGCAAAUGAACGGCCUGGGGCGGACAGACCAGCCAGG----------CGAAGGCCAAGGCCCCGCCCAGCACUUAGAGGCUG ..(((....)))....(((((((((.(((...((((((........(((.(((..(.....).)))))).((----------(....)))...))))))....)))))))))))).. ( -43.80) >consensus GCCAACGCAGCAAGUUGUCUCCAAGGUAUUUUUGCACCAGAUGAAUGGCCAGGGGCUCUGGGCCCGGUCAAC_U__GGUGAGAAAAUACCAAUGCUGCUUU_____________CUG ......((((((..(((....)))(((((((((.((((.......(((((..((((.....)))))))))......)))).)))))))))..))))))................... (-23.14 = -26.42 + 3.28)


| Location | 8,400,702 – 8,400,803 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.99 |
| Mean single sequence MFE | -38.86 |
| Consensus MFE | -20.00 |
| Energy contribution | -22.25 |
| Covariance contribution | 2.26 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8400702 101 - 27905053 CAA-------------AAAGUGCCAUUGGAAUUUUCUCACC--A-GUUGACCGGGCGCAGAGCCCCUGGUCAUUCAUCUGGUGCAAAAAUACCUUGGAGACAACUUGCUGCGUUGGC ...-------------.....((((..((.(((((..((((--(-(.((((((((.((...)).)))))))).....))))))..))))).))(((....)))..........)))) ( -35.60) >DroSec_CAF1 62553 102 - 1 CAG-------------AAAGCAGCAUUGGUAUUUGCUUUCC--UUCCUGACCGGGCCCAAAGCUCCUGGCCAUUCAUCUGGUGCAAAAAUACCUUGGAGACAACUUGCUGCGUUGGC (((-------------...((((((..(((((((((...((--....(((...((((..........))))..)))...)).)))..))))))(((....)))..)))))).))).. ( -32.40) >DroSim_CAF1 61720 101 - 1 CAG-------------AAAGCAGCAUUGGUAUUUCCUCACC--A-GUUGACCGGGCCCAGAGCCCCUGGCCAUUCAUCUGGUGCAAAAAUACCUUGGAGACAACUUGCUGCGUUGGC (((-------------...((((((..(((((((...((((--(-(.(((...((((.((.....))))))..))).))))))...)))))))(((....)))..)))))).))).. ( -42.40) >DroEre_CAF1 64520 101 - 1 CAG-------------AAAGCAGCAUUGGUAUUUUCUCACC--A-GUUGACCGGGCCCAGAGGACCUGGCCAUUCAUCUGGUGCAAAAAUACCUUGGAGACAACUUGCUGCGGUGGC ...-------------...((((((..((((((((..((((--(-(.(((...((((.((.....))))))..))).))))))..))))))))(((....)))..))))))...... ( -43.60) >DroYak_CAF1 64347 103 - 1 CAG-------------AAAGCAGCAUUGGUAUUUUCCCACCCAG-GUUGACCGGGCCCAGAGCCCCUGGCCAUUCAUCUGGUGCAAAAAUACCUUGGAGACAACUUGCUGCGGUGGC ...-------------...((((((..((((((((..((((.((-((.((...((((.((.....))))))..))))))))))..))))))))(((....)))..))))))...... ( -40.00) >DroPer_CAF1 65175 107 - 1 CAGCCUCUAAGUGCUGGGCGGGGCCUUGGCCUUCG----------CCUGGCUGGUCUGUCCGCCCCAGGCCGUUCAUUUGCCCCAAAGACACCUCAGAGACACAUUGCGGCGGCAGU ..(((.......((..((((((((....))).)))----------))..)).((((((.......))))))........(((.(((..................))).))))))... ( -39.17) >consensus CAG_____________AAAGCAGCAUUGGUAUUUUCUCACC__A_GUUGACCGGGCCCAGAGCCCCUGGCCAUUCAUCUGGUGCAAAAAUACCUUGGAGACAACUUGCUGCGGUGGC ...................((((((..(((((((...((((......((.(((((.........))))).)).......))))...)))))))(((....)))..))))))...... (-20.00 = -22.25 + 2.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:48 2006