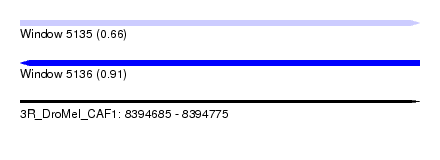
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,394,685 – 8,394,775 |
| Length | 90 |
| Max. P | 0.912035 |

| Location | 8,394,685 – 8,394,775 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 80.89 |
| Mean single sequence MFE | -21.80 |
| Consensus MFE | -15.49 |
| Energy contribution | -16.43 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8394685 90 + 27905053 GCCACACA-CACACGA-----UUUUUC---CGCUUUCCACGCUGGCUUUUCGCAUUCCACGCGUGGCCAGCGAUUUUCGCCGCUCUGCGUUU-UGAUUAU ........-.......-----......---.........((((((((...(((.......))).)))))))).....(((......)))...-....... ( -21.70) >DroSec_CAF1 56558 90 + 1 GCCACACA-CACAUGA-----AUUUUC---CGCUUUCCACGCUGGCUUUUCGCAUUCCACGCGUGGCCAGCGAUUUUCGCCGCUCUGCGUUU-UGAUUAU ........-.......-----......---.........((((((((...(((.......))).)))))))).....(((......)))...-....... ( -21.70) >DroSim_CAF1 55865 81 + 1 ----------ACAUGA-----AUUUUC---CGCUUUCCACGCUGGCUUUUCGCAUUCCACGCGUGGCCAGCGAUUUUCGCCGCUCUGCGUUU-UGAUUAU ----------......-----......---.........((((((((...(((.......))).)))))))).....(((......)))...-....... ( -21.70) >DroEre_CAF1 58427 91 + 1 GCCACACA---UACGA-----AUUUUUCCACGCUUUCCACGCUGGCUUCUCGCAUUCCACGCGUGGCCAGCGAUUUUCCCUGCUCUGCGUUU-UGAUUAU ........---.....-----........((((....((((((((((...(((.......))).))))))))........))....))))..-....... ( -21.40) >DroYak_CAF1 58172 78 + 1 GCCACACA---CACGA-----AUUUUUCCACGCUUUCCACGCUGGCUUUUCGCAUUCCACGCGUGGCC-------------GCUCUGCGUUU-UGAUUAU (((((...---...((-----(.....(((.((.......)))))...)))((.......)))))))(-------------((...)))...-....... ( -15.80) >DroAna_CAF1 58933 95 + 1 GCCGCACAGCGUUCGACUGAGAAUUUC---CACUUGCCA--CUGGCUUUUCGCAUUCCACGCGUGGCCGGCGAUUUUCGCCGCUUUGCGUUUCUGAUUAU ..((((.(((((((......))))...---.........--..((((...(((.......))).))))((((.....))))))).))))........... ( -28.50) >consensus GCCACACA__ACACGA_____AUUUUC___CGCUUUCCACGCUGGCUUUUCGCAUUCCACGCGUGGCCAGCGAUUUUCGCCGCUCUGCGUUU_UGAUUAU .......................................((((((((...(((.......))).))))))))........(((...)))........... (-15.49 = -16.43 + 0.95)


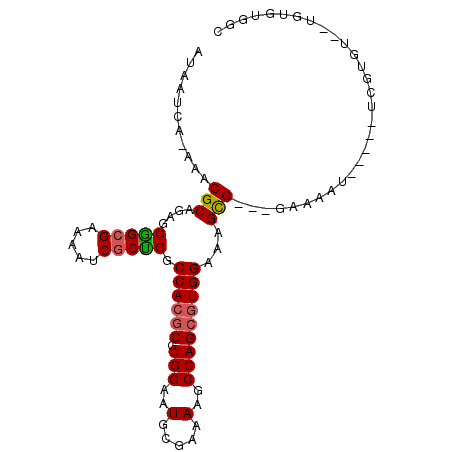
| Location | 8,394,685 – 8,394,775 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 80.89 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -20.03 |
| Energy contribution | -20.33 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8394685 90 - 27905053 AUAAUCA-AAACGCAGAGCGGCGAAAAUCGCUGGCCACGCGUGGAAUGCGAAAAGCCAGCGUGGAAAGCG---GAAAAA-----UCGUGUG-UGUGUGGC .......-..(((((..(((.(((...(((((..((((((.(((..(.....)..)))))))))..))))---).....-----)))))).-)))))... ( -31.00) >DroSec_CAF1 56558 90 - 1 AUAAUCA-AAACGCAGAGCGGCGAAAAUCGCUGGCCACGCGUGGAAUGCGAAAAGCCAGCGUGGAAAGCG---GAAAAU-----UCAUGUG-UGUGUGGC .......-..(((((..(((..(((..(((((..((((((.(((..(.....)..)))))))))..))))---)....)-----)).))).-)))))... ( -29.70) >DroSim_CAF1 55865 81 - 1 AUAAUCA-AAACGCAGAGCGGCGAAAAUCGCUGGCCACGCGUGGAAUGCGAAAAGCCAGCGUGGAAAGCG---GAAAAU-----UCAUGU---------- .......-...(((......)))....(((((..((((((.(((..(.....)..)))))))))..))))---).....-----......---------- ( -25.90) >DroEre_CAF1 58427 91 - 1 AUAAUCA-AAACGCAGAGCAGGGAAAAUCGCUGGCCACGCGUGGAAUGCGAGAAGCCAGCGUGGAAAGCGUGGAAAAAU-----UCGUA---UGUGUGGC .......-..(((((..((..(......((((..((((((.(((...........)))))))))..))))........)-----..)).---)))))... ( -26.34) >DroYak_CAF1 58172 78 - 1 AUAAUCA-AAACGCAGAGC-------------GGCCACGCGUGGAAUGCGAAAAGCCAGCGUGGAAAGCGUGGAAAAAU-----UCGUG---UGUGUGGC .......-...(((...))-------------)(((((((.....(((((((...(((.(((.....)))))).....)-----)))))---)))))))) ( -22.60) >DroAna_CAF1 58933 95 - 1 AUAAUCAGAAACGCAAAGCGGCGAAAAUCGCCGGCCACGCGUGGAAUGCGAAAAGCCAG--UGGCAAGUG---GAAAUUCUCAGUCGAACGCUGUGCGGC ...........((((.(((((((.....))))..((((((((...)))).....(((..--.)))..)))---)................))).)))).. ( -29.60) >consensus AUAAUCA_AAACGCAGAGCGGCGAAAAUCGCUGGCCACGCGUGGAAUGCGAAAAGCCAGCGUGGAAAGCG___GAAAAU_____UCGUGU__UGUGUGGC ...........(((....(((((.....))))).((((((.(((..(.....)..)))))))))...))).............................. (-20.03 = -20.33 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:46 2006