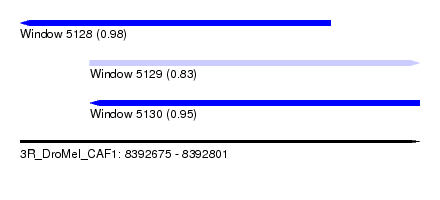
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,392,675 – 8,392,801 |
| Length | 126 |
| Max. P | 0.976981 |

| Location | 8,392,675 – 8,392,773 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.77 |
| Mean single sequence MFE | -36.95 |
| Consensus MFE | -26.84 |
| Energy contribution | -27.32 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
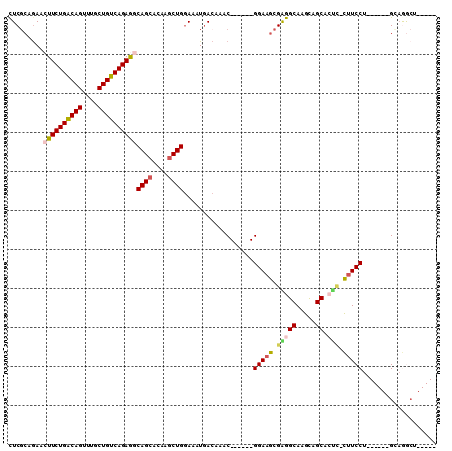
>3R_DroMel_CAF1 8392675 98 - 27905053 CUCGCAGAACUUCUGACAGUUUGCUGUCAGAGGCAGCACAAGCUGGAAAUGACAAACGGAAACGGAAGCGAGGCAAGCAGCACUC-CUUCCU------GCAGGCU----- (((((....((((((((((....))))))))))((((....))))...........((....))...)))))((..((((.....-....))------))..)).----- ( -39.30) >DroVir_CAF1 69440 92 - 1 CUCGCCAAACUUCUGGCAGUUUGCUGUCAGAAACAGCAAAUGCUG-AAAUCAGAAAC------GGAAGCGGGGCAAGUGGCAGCUCUUUCCU------GGAAACA----- ....(((...(((((((((....))))))))).((((....))))-...........------((((..(((((........))))))))))------)).....----- ( -29.50) >DroSim_CAF1 53831 98 - 1 CUCGCAGAACUUCUGACAGUUUGCUGUCAGAGGCAGCACAAGCUGGAAAUGACAAACGGAAACGGAAGCGGGGCAAGCAGCACUC-CUUCCU------GCAGGCU----- (((((....((((((((((....))))))))))((((....))))...........((....))...)))))((..((((.....-....))------))..)).----- ( -38.40) >DroYak_CAF1 56162 98 - 1 CUCGCAGAACUUCUGACAGUUUGCUGUCAGAGGCAGCACAAGCUGGAAAUGACAAACGGAAACGGAAGCGGGGCAGGCAGCACUC-CUUCCU------GCAGGCU----- (((((....((((((((((....))))))))))((((....))))...........((....))...)))))(((((.((.....-)).)))------)).....----- ( -40.90) >DroAna_CAF1 56723 90 - 1 UUCGCCGAACUUCUGACAGUUUGCUGUCAGAGGCAGCGCAGACUGGAAAUGACAAAC------GGAAGCGAGGCAGGCAGCACUA-CGUCCU------GGCGG------- ..(((((..((((((((((....))))))))))(((......)))............------(((....((((.....)).)).-..))))------)))).------- ( -30.50) >DroPer_CAF1 55855 103 - 1 CUCUGCGAAGUUCUGACAGUUUGCUGUCAGAGGCAGCGAAGGCUGGAAAUGACAAAC------GGAAGCGAGGCAGGCAGCAGCU-CUUCCUCUUCCUGGAAACAGGAAC .((((.....(((((((((....))))))))).((((....))))...........)------)))...((((.((((....)).-)).))))((((((....)))))). ( -43.10) >consensus CUCGCAGAACUUCUGACAGUUUGCUGUCAGAGGCAGCACAAGCUGGAAAUGACAAAC______GGAAGCGAGGCAAGCAGCACUC_CUUCCU______GCAGGCU_____ .........((((((((((....))))))))))((((....))))..................(((((.(((((.....)).))).)))))................... (-26.84 = -27.32 + 0.47)

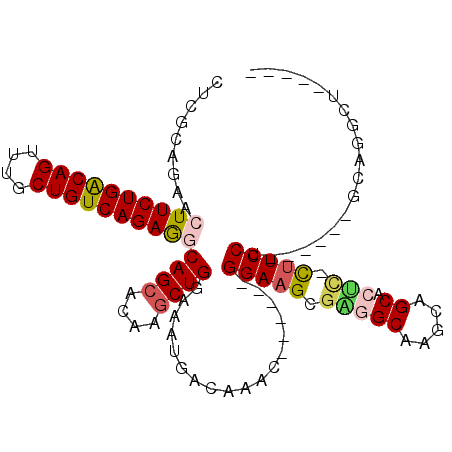
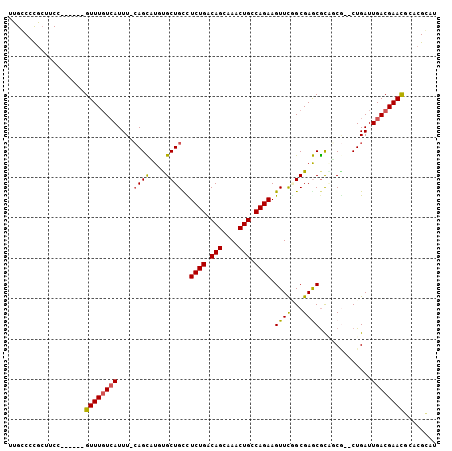
| Location | 8,392,697 – 8,392,801 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 79.49 |
| Mean single sequence MFE | -33.93 |
| Consensus MFE | -18.67 |
| Energy contribution | -18.33 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8392697 104 + 27905053 UUGCCUCGCUUCCGUUUCCGUUUGUCAUUUCCAGCUUGUGCUGCCUCUGACAGCAAACUGUCAGAAGUUCUGCGAGCGUUGCG--CUGAUUGACGAACCCACGGAA ..........(((((....((((((((....(((((((((..((.((((((((....)))))))).))..)))))((...)))--)))..))))))))..))))). ( -36.70) >DroVir_CAF1 69463 97 + 1 UUGCCCCGCUUCC------GUUUCUGAUUU-CAGCAUUUGCUGUUUCUGACAGCAAACUGCCAGAAGUUUGGCGAGCGCAUCG--CUGAUUGACGAACGCACGCUU (((((..(((((.------.....((....-))((((((((((((...))))))))).)))..)))))..)))))(((..(((--(.....).))).)))...... ( -29.10) >DroGri_CAF1 62090 97 + 1 UUGGCCUGCUUCC------GUUUGUCAUUU-CAGUAUUUGCUGUUUCUGACAGCAAGCUGCCAGAAGUUUAGCGAACGCAUCU--CUGAUUGACGAAUGUACGCUU .......((...(------((((((((..(-(((..(((((((((((((.(((....))).))))))..))))))).......--)))).)))))))))...)).. ( -29.70) >DroEre_CAF1 56361 104 + 1 CUGCCCCACUUCCGUUUCCGUUUGUCAUUUCCAGCUUGCGCUGCCUCUGACAGCAAACUGUCAGAAGUUCUGCGAGCGUUGCG--CUGAUUGACGAACCCACGGAA ..........(((((....((((((((....((((..((((((((((((((((....))))))).))....)).)))))...)--)))..))))))))..))))). ( -39.60) >DroMoj_CAF1 64924 99 + 1 UUGGCCCGAUUCC------GUUUGUCAUUU-GAGCAUUUGCUGCUUCUGACAGCCAGCUGCCAGAAGUUUGGCGAGCGCGCCAACCUGAUUGACGAACGCACGCUU ..(((.......(------((((((((.((-.(((....)))(((((((.(((....))).)))))))((((((....))))))...)).)))))))))...))). ( -34.00) >DroAna_CAF1 56743 98 + 1 CUGCCUCGCUUCC------GUUUGUCAUUUCCAGUCUGCGCUGCCUCUGACAGCAAACUGUCAGAAGUUCGGCGAACAUGGCG--CUGAUUGACGAACGCACGGAU ..........(((------((.(((......(((((.(((((...((((((((....)))))))).((((...))))..))))--).)))))....))).))))). ( -34.50) >consensus UUGCCCCGCUUCC______GUUUGUCAUUU_CAGCAUGUGCUGCCUCUGACAGCAAACUGCCAGAAGUUCGGCGAGCGCAGCG__CUGAUUGACGAACGCACGCAU ...................((((((((....((((....))))..((((.(((....))).)))).((((...)))).............))))))))........ (-18.67 = -18.33 + -0.33)

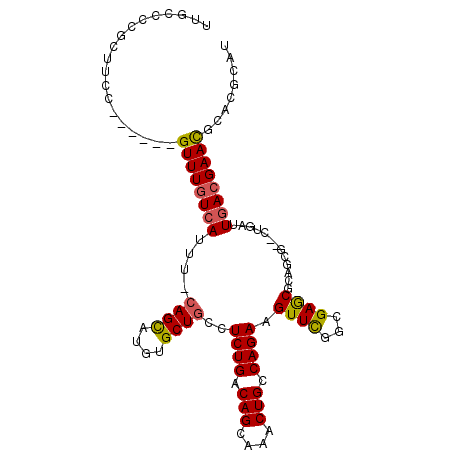
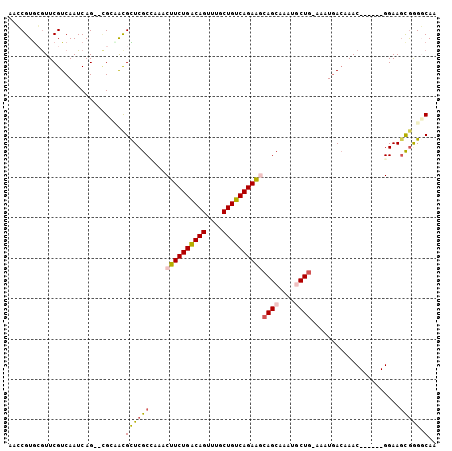
| Location | 8,392,697 – 8,392,801 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 79.49 |
| Mean single sequence MFE | -35.22 |
| Consensus MFE | -22.75 |
| Energy contribution | -22.83 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8392697 104 - 27905053 UUCCGUGGGUUCGUCAAUCAG--CGCAACGCUCGCAGAACUUCUGACAGUUUGCUGUCAGAGGCAGCACAAGCUGGAAAUGACAAACGGAAACGGAAGCGAGGCAA ....((..((((((......)--)).))).(((((....((((((((((....))))))))))((((....))))...........((....))...))))))).. ( -36.90) >DroVir_CAF1 69463 97 - 1 AAGCGUGCGUUCGUCAAUCAG--CGAUGCGCUCGCCAAACUUCUGGCAGUUUGCUGUCAGAAACAGCAAAUGCUG-AAAUCAGAAAC------GGAAGCGGGGCAA ..(.((((((.(((......)--)))))))).)(((...((((((...(((((((((.....))))))))).(((-....)))...)------)))))...))).. ( -36.40) >DroGri_CAF1 62090 97 - 1 AAGCGUACAUUCGUCAAUCAG--AGAUGCGUUCGCUAAACUUCUGGCAGCUUGCUGUCAGAAACAGCAAAUACUG-AAAUGACAAAC------GGAAGCAGGCCAA ..((...(.((.((((.((((--.((.....))(((....(((((((((....)))))))))..))).....)))-)..)))).)).------)...))....... ( -26.00) >DroEre_CAF1 56361 104 - 1 UUCCGUGGGUUCGUCAAUCAG--CGCAACGCUCGCAGAACUUCUGACAGUUUGCUGUCAGAGGCAGCGCAAGCUGGAAAUGACAAACGGAAACGGAAGUGGGGCAG ((((((..(((..((.....(--((.......)))....((((((((((....))))))))))((((....))))))...)))..))))))((....))....... ( -40.40) >DroMoj_CAF1 64924 99 - 1 AAGCGUGCGUUCGUCAAUCAGGUUGGCGCGCUCGCCAAACUUCUGGCAGCUGGCUGUCAGAAGCAGCAAAUGCUC-AAAUGACAAAC------GGAAUCGGGCCAA ..((.((((((.((((......((((((....)))))).((((((((((....)))))))))).(((....))).-...)))).)))------)....)).))... ( -34.50) >DroAna_CAF1 56743 98 - 1 AUCCGUGCGUUCGUCAAUCAG--CGCCAUGUUCGCCGAACUUCUGACAGUUUGCUGUCAGAGGCAGCGCAGACUGGAAAUGACAAAC------GGAAGCGAGGCAG ..((.(((.(((((...(((.--..(((.((..(((...((((((((((....))))))))))..).))..)))))...)))...))------))).))).))... ( -37.10) >consensus AACCGUGCGUUCGUCAAUCAG__CGCAACGCUCGCCAAACUUCUGACAGUUUGCUGUCAGAAGCAGCAAAUGCUG_AAAUGACAAAC______GGAAGCGGGGCAA .............................((((((....((((((((((....))))))))))((((....))))......................))))).).. (-22.75 = -22.83 + 0.09)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:41 2006