
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,370,855 – 8,371,014 |
| Length | 159 |
| Max. P | 0.997930 |

| Location | 8,370,855 – 8,370,945 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 77.33 |
| Mean single sequence MFE | -18.96 |
| Consensus MFE | -6.23 |
| Energy contribution | -5.95 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.33 |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.997930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8370855 90 + 27905053 GUAACGUU---------CUUUGCACACAAAAACCGAAAUAGCAACAGAAUCAGAUUCAGAAUCCGAAUCCGAAUCUGAAUCUGAAUCUGAAUCAGAGUC ......((---------((((((.................))))((((.(((((((((((.((.......)).))))))))))).))))....)))).. ( -23.93) >DroSec_CAF1 32778 80 + 1 GUAACGUU---------CUGUGCACACAAAAACCGAAAUAGCAACAGAAUCCGAUUCAGAAUCCGAAUCUGAAUC----------UUUGAAUCAGAGUC .....(((---------((((((.................)).)))))))..((((((((.......))))))))----------.............. ( -16.43) >DroSim_CAF1 31607 80 + 1 GUAACGUU---------CUGUGCACACAAAAACCGAAAUAGCAACAGAAUCCGAUUCAGAAUCCGAAUCUGAAUC----------UCUGAAUCAGAGUC .....(((---------((((((.................)).)))))))..((((((((.......))))))))----------((((...))))... ( -17.73) >DroEre_CAF1 33853 86 + 1 -UAACGGUCUUUCCAGACUUUGCACACAAAAACAAAAAUAGAAACAAACUCAGAUUCUGAAUCUGAAUCAGAAU------------CAGAAUCAGAGUC -....(((((....)))))............................((((.((((((((.((((...)))).)------------))))))).)))). ( -21.30) >DroYak_CAF1 33554 86 + 1 -UUACGGUCUUUGCAGUCUUUGCACACAACAACAAAAAUAGAAACAAAUUCACAUUCAGAUUCUGAAUCUGAAU------------CUGAAUCAGAGUC -..........(((((...))))).......................((((..((((((((((.......))))------------))))))..)))). ( -15.40) >consensus GUAACGUU_________CUUUGCACACAAAAACCGAAAUAGCAACAGAAUCAGAUUCAGAAUCCGAAUCUGAAUC__________UCUGAAUCAGAGUC .................................................((.(((((((...........................))))))).))... ( -6.23 = -5.95 + -0.28)



| Location | 8,370,855 – 8,370,945 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 77.33 |
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -8.14 |
| Energy contribution | -8.10 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.33 |
| SVM decision value | 2.94 |
| SVM RNA-class probability | 0.997821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8370855 90 - 27905053 GACUCUGAUUCAGAUUCAGAUUCAGAUUCGGAUUCGGAUUCUGAAUCUGAUUCUGUUGCUAUUUCGGUUUUUGUGUGCAAAG---------AACGUUAC (((.((((...((...((((.(((((((((((.......))))))))))).))))...))...))))((((((....)))))---------)..))).. ( -29.20) >DroSec_CAF1 32778 80 - 1 GACUCUGAUUCAAA----------GAUUCAGAUUCGGAUUCUGAAUCGGAUUCUGUUGCUAUUUCGGUUUUUGUGUGCACAG---------AACGUUAC ((((((((((((..----------(((((......))))).)))))))))((((((.(((((..........))).))))))---------)).))).. ( -19.80) >DroSim_CAF1 31607 80 - 1 GACUCUGAUUCAGA----------GAUUCAGAUUCGGAUUCUGAAUCGGAUUCUGUUGCUAUUUCGGUUUUUGUGUGCACAG---------AACGUUAC ((((((((((((((----------(.(((......)))))))))))))))((((((.(((((..........))).))))))---------)).))).. ( -23.90) >DroEre_CAF1 33853 86 - 1 GACUCUGAUUCUG------------AUUCUGAUUCAGAUUCAGAAUCUGAGUUUGUUUCUAUUUUUGUUUUUGUGUGCAAAGUCUGGAAAGACCGUUA- (((((.(((((((------------(.((((...)))).)))))))).)))))..........(((((........)))))((((....)))).....- ( -27.40) >DroYak_CAF1 33554 86 - 1 GACUCUGAUUCAG------------AUUCAGAUUCAGAAUCUGAAUGUGAAUUUGUUUCUAUUUUUGUUGUUGUGUGCAAAGACUGCAAAGACCGUAA- ......((..(((------------(((((.((((((...)))))).))))))))..))..(((((((.(((.........))).)))))))......- ( -22.80) >consensus GACUCUGAUUCAGA__________GAUUCAGAUUCGGAUUCUGAAUCUGAUUCUGUUGCUAUUUCGGUUUUUGUGUGCAAAG_________AACGUUAC ........................((((((((.......)))))))).................................................... ( -8.14 = -8.10 + -0.04)



| Location | 8,370,868 – 8,370,978 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 86.90 |
| Mean single sequence MFE | -22.46 |
| Consensus MFE | -11.30 |
| Energy contribution | -11.50 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.54 |
| Structure conservation index | 0.50 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8370868 110 + 27905053 CACACAAAAACCGAAAUAGCAACAGAAUCAGAUUCAGAAUCCGAAUCCGAAUCUGAAUCUGAAUCUGAAUCAGAGUCCCAGUGAACUCGAAACCAUUCGAAAGUUUCCAG ...........((((.......((((.(((((((((((.((.......)).))))))))))).)))).....((((........)))).......))))........... ( -28.50) >DroSec_CAF1 32791 100 + 1 CACACAAAAACCGAAAUAGCAACAGAAUCCGAUUCAGAAUCCGAAUCUGAAUC----------UUUGAAUCAGAGUCCCAGUGAACUCGAAACCAUUCGAAAGUUUCCAG ............(((((.............((((((((.......))))))))----------(((((((..((((........))))......))))))).)))))... ( -17.70) >DroSim_CAF1 31620 100 + 1 CACACAAAAACCGAAAUAGCAACAGAAUCCGAUUCAGAAUCCGAAUCUGAAUC----------UCUGAAUCAGAGUCCCAGUGAACUCGAAACCAUUCGAAAGUUUCCAG ...........((((.......((((....((((((((.......))))))))----------)))).....((((........)))).......))))........... ( -18.20) >DroEre_CAF1 33874 98 + 1 CACACAAAAACAAAAAUAGAAACAAACUCAGAUUCUGAAUCUGAAUCAGAAU------------CAGAAUCAGAGUCCCACUGAACUCGAAGCCAUUCGAAAGUUUCCAG ..................(((((..((((.((((((((.((((...)))).)------------))))))).))))..........(((((....)))))..)))))... ( -26.80) >DroYak_CAF1 33575 98 + 1 CACACAACAACAAAAAUAGAAACAAAUUCACAUUCAGAUUCUGAAUCUGAAU------------CUGAAUCAGAGUCCCACUGAACUCGAAACCAUUCGAAAGUUUCCAG ..................(((((..((((..((((((((((.......))))------------))))))..))))..........(((((....)))))..)))))... ( -21.10) >consensus CACACAAAAACCGAAAUAGCAACAGAAUCAGAUUCAGAAUCCGAAUCUGAAUC__________UCUGAAUCAGAGUCCCAGUGAACUCGAAACCAUUCGAAAGUUUCCAG ..............................((((((((.......))))))))...................((((........))))(((((.........)))))... (-11.30 = -11.50 + 0.20)


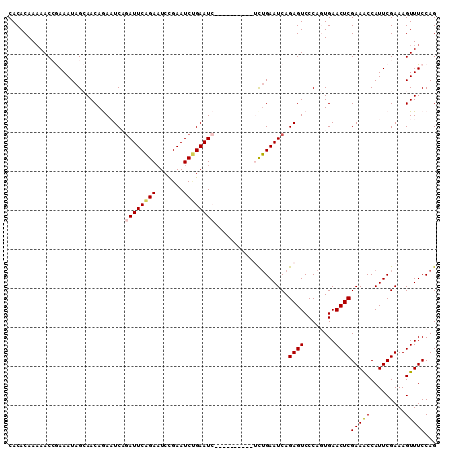
| Location | 8,370,868 – 8,370,978 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 86.90 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -20.52 |
| Energy contribution | -21.48 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8370868 110 - 27905053 CUGGAAACUUUCGAAUGGUUUCGAGUUCACUGGGACUCUGAUUCAGAUUCAGAUUCAGAUUCGGAUUCGGAUUCUGAAUCUGAUUCUGUUGCUAUUUCGGUUUUUGUGUG ..(((((((...(((((((...((((((....)))))).....((((.(((((((((((((((....)))).))))))))))).))))..))))))).)))))))..... ( -39.30) >DroSec_CAF1 32791 100 - 1 CUGGAAACUUUCGAAUGGUUUCGAGUUCACUGGGACUCUGAUUCAAA----------GAUUCAGAUUCGGAUUCUGAAUCGGAUUCUGUUGCUAUUUCGGUUUUUGUGUG ..(((((((...(((((((..(((((((....)))))).((.((...----------((((((((.......)))))))).)).)).)..))))))).)))))))..... ( -29.60) >DroSim_CAF1 31620 100 - 1 CUGGAAACUUUCGAAUGGUUUCGAGUUCACUGGGACUCUGAUUCAGA----------GAUUCAGAUUCGGAUUCUGAAUCGGAUUCUGUUGCUAUUUCGGUUUUUGUGUG ..(((((((...(((((((..(((((((....)))))).((.((...----------((((((((.......)))))))).)).)).)..))))))).)))))))..... ( -29.60) >DroEre_CAF1 33874 98 - 1 CUGGAAACUUUCGAAUGGCUUCGAGUUCAGUGGGACUCUGAUUCUG------------AUUCUGAUUCAGAUUCAGAAUCUGAGUUUGUUUCUAUUUUUGUUUUUGUGUG .(((((((.((((((....)))))).......((((((.(((((((------------(.((((...)))).)))))))).)))))))))))))................ ( -31.80) >DroYak_CAF1 33575 98 - 1 CUGGAAACUUUCGAAUGGUUUCGAGUUCAGUGGGACUCUGAUUCAG------------AUUCAGAUUCAGAAUCUGAAUGUGAAUUUGUUUCUAUUUUUGUUGUUGUGUG ...((((((.......))))))((((((....)))))).((..(((------------(((((.((((((...)))))).))))))))..)).................. ( -29.30) >consensus CUGGAAACUUUCGAAUGGUUUCGAGUUCACUGGGACUCUGAUUCAGA__________GAUUCAGAUUCGGAUUCUGAAUCUGAUUCUGUUGCUAUUUCGGUUUUUGUGUG ..(((((((...(((((((..(((((((....))))))...................((((((((.......)))))))).......)..))))))).)))))))..... (-20.52 = -21.48 + 0.96)


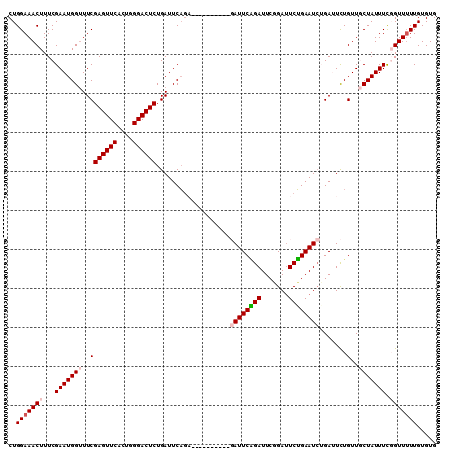
| Location | 8,370,905 – 8,371,014 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 89.71 |
| Mean single sequence MFE | -28.73 |
| Consensus MFE | -23.03 |
| Energy contribution | -23.54 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8370905 109 - 27905053 AUACAUGCGUAUGCUCGUGUAUGCAUGUUUUAUUUGCUGGAAACUUUCGAAUGGUUUCGAGUUCACUGGGACUCUGAUUCAGAUUCAGAUUCAGAUUCGGAUUCGGAUU ..(((((((((((....))))))))))).(((((((..(....)...))))))).(((((((((.(((((..(((((.......))))))))))....))))))))).. ( -32.50) >DroSim_CAF1 31657 99 - 1 AUACAUGCGUAUGCUCGUGUAUGCAUGUUUUAUUUGCUGGAAACUUUCGAAUGGUUUCGAGUUCACUGGGACUCUGAUUCAGA----------GAUUCAGAUUCGGAUU ..(((((((((((....))))))))))).(((((((..(....)...))))))).(((((((...(((((.(((((...))))----------).)))))))))))).. ( -30.80) >DroEre_CAF1 33911 97 - 1 AUACAUGCGUAUGCUCGUGUAUGGAUGUUUUAUUUGCUGGAAACUUUCGAAUGGCUUCGAGUUCAGUGGGACUCUGAUUCUG------------AUUCUGAUUCAGAUU ..((((.((((((....)))))).))))....((..(((((....((((((....)))))))))))..))..(((((.((..------------.....)).))))).. ( -24.60) >DroYak_CAF1 33612 97 - 1 AUACAUGCGUAUGCUCGUGUAUGGAUGUUUUAUUUGCUGGAAACUUUCGAAUGGUUUCGAGUUCAGUGGGACUCUGAUUCAG------------AUUCAGAUUCAGAAU (((((((.(....).)))))))....((((((((.(((.((((((.......)))))).)))..))))))))(((((.((..------------.....)).))))).. ( -27.00) >consensus AUACAUGCGUAUGCUCGUGUAUGCAUGUUUUAUUUGCUGGAAACUUUCGAAUGGUUUCGAGUUCACUGGGACUCUGAUUCAG____________AUUCAGAUUCAGAUU (((((((.(....).)))))))....((((((((.(((.((((((.......)))))).)))..))))))))(((((.((...................)).))))).. (-23.03 = -23.54 + 0.50)

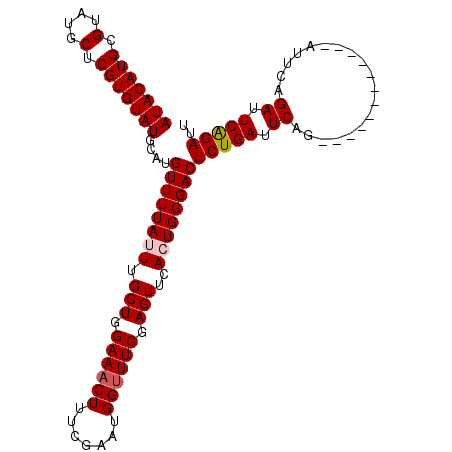

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:33 2006