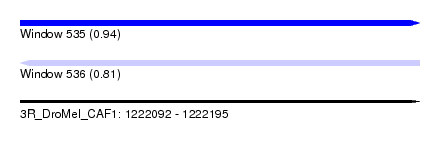
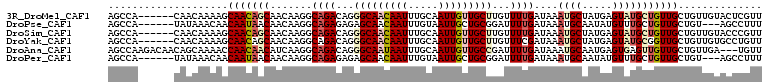
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,222,092 – 1,222,195 |
| Length | 103 |
| Max. P | 0.940501 |

| Location | 1,222,092 – 1,222,195 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 81.42 |
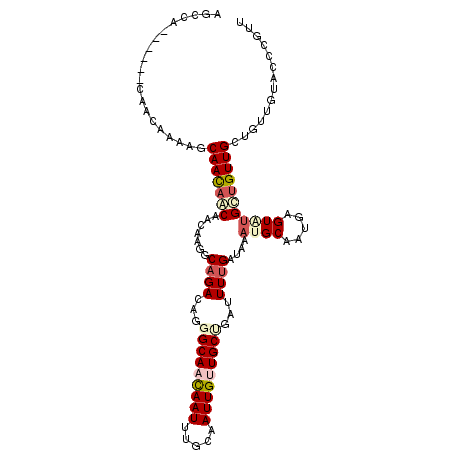
| Mean single sequence MFE | -28.42 |
| Consensus MFE | -20.03 |
| Energy contribution | -20.45 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
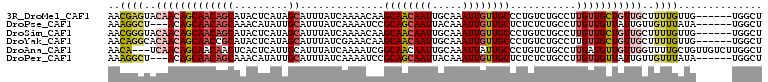
>3R_DroMel_CAF1 1222092 103 + 27905053 AACGAGUACAACAGCAACAGCAUACUCAUAGCAUUUAUCAAAACAAGCAACAAUUGCAAAUUGUUGCCCUGUCUGCCUUGUUGCUGUUGCUUUUGUUG------UGGCU ....((((((((((((((((((....((..(((.........(((.((((((((.....))))))))..))).)))..)).)))))))))...)))))------).))) ( -32.30) >DroPse_CAF1 2337 100 + 1 AAAGGCU---ACAGCAACAGCAAACAUAUUGCAUUUAUCAAAAUCCGCAGCAAUUACAAAUUGUUGCUCUCUCUGCCUUGUUGUUAUUGUUGUUUAUA------UGGCU ...((((---(.(((((((((((.....))))..............((((((((.....))))))))....................)))))))....------))))) ( -21.90) >DroSim_CAF1 2038 103 + 1 AACGGGUACAACAGCAACAGCAUACUCAUAGCAUUUAUCAAAACAAGCAACAAUUGCAAAUUGUUGCCCUGUCUGCCUUGUUGCUGUUGCUUUUGUUG------UGGCU ...((.((((((((((((((((....((..(((.........(((.((((((((.....))))))))..))).)))..)).)))))))))...)))))------)).)) ( -34.50) >DroYak_CAF1 2064 103 + 1 AACAGGCACAACAGCAACCGCAUACUCAUAGCAUUUAUCGAAACAAGCAACAAUUGCAAAUUGUUGCCCUGUCUGCCUUGUUGCUGUUGCUUUUGUUG------UGGCU ...((.((((((((((((.(((....((..(((......((.....((((((((.....))))))))....)))))..)).))).)))))...)))))------)).)) ( -32.20) >DroAna_CAF1 2052 106 + 1 AACA---UCAACAGCAACAACUCACUCAUUGCAUUUAUCAAAAUCGGCAACAAUUGCAAAUUAUUGCCCUGUCUGCCUUGAUGUUGUUGGUUUUGCUGUUGUCUUGGCU ..((---.(((((((((.(((.((......(((..((((((....(((((((...((((....))))..))).)))))))))).)))))))))))))))))...))... ( -27.70) >DroPer_CAF1 4212 100 + 1 AAAGGCU---ACAGCAACAGCAAACAUAUUGCAUUUAUCAAAAUCCGCAGCAAUUACAAAUUGUUGCUCUCUCUGCCUUGUUGUUAUUGUUGUUUAUA------UGGCU ...((((---(.(((((((((((.....))))..............((((((((.....))))))))....................)))))))....------))))) ( -21.90) >consensus AACGGGUACAACAGCAACAGCAUACUCAUAGCAUUUAUCAAAACAAGCAACAAUUGCAAAUUGUUGCCCUGUCUGCCUUGUUGCUGUUGCUUUUGUUG______UGGCU ..((((..(((((((((((((.........))..............((((((((.....))))))))...........)))))))))))..)))).............. (-20.03 = -20.45 + 0.42)
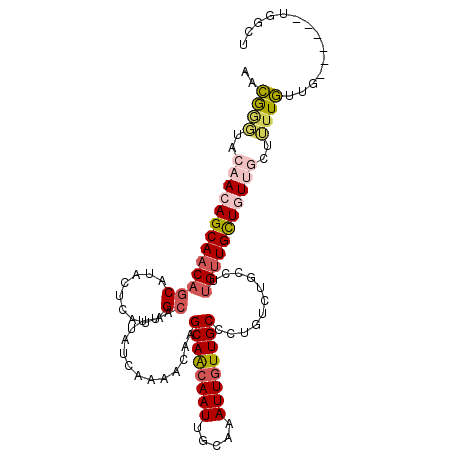
| Location | 1,222,092 – 1,222,195 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -15.03 |
| Energy contribution | -16.17 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1222092 103 - 27905053 AGCCA------CAACAAAAGCAACAGCAACAAGGCAGACAGGGCAACAAUUUGCAAUUGUUGCUUGUUUUGAUAAAUGCUAUGAGUAUGCUGUUGCUGUUGUACUCGUU ....(------(((((...((((((((..(((((((.....(((((((((.....))))))))))))))))....((((.....))))))))))))))))))....... ( -35.60) >DroPse_CAF1 2337 100 - 1 AGCCA------UAUAAACAACAAUAACAACAAGGCAGAGAGAGCAACAAUUUGUAAUUGCUGCGGAUUUUGAUAAAUGCAAUAUGUUUGCUGUUGCUGU---AGCCUUU .....------...................(((((....(.((((((((((((((.....)))))))).........((((.....)))).)))))).)---.))))). ( -21.50) >DroSim_CAF1 2038 103 - 1 AGCCA------CAACAAAAGCAACAGCAACAAGGCAGACAGGGCAACAAUUUGCAAUUGUUGCUUGUUUUGAUAAAUGCUAUGAGUAUGCUGUUGCUGUUGUACCCGUU ....(------(((((...((((((((..(((((((.....(((((((((.....))))))))))))))))....((((.....))))))))))))))))))....... ( -35.60) >DroYak_CAF1 2064 103 - 1 AGCCA------CAACAAAAGCAACAGCAACAAGGCAGACAGGGCAACAAUUUGCAAUUGUUGCUUGUUUCGAUAAAUGCUAUGAGUAUGCGGUUGCUGUUGUGCCUGUU .....------.((((...(((((((((((...(((((((((.(((((((.....))))))))))))))......((((.....)))))).)))))))))))...)))) ( -35.70) >DroAna_CAF1 2052 106 - 1 AGCCAAGACAACAGCAAAACCAACAACAUCAAGGCAGACAGGGCAAUAAUUUGCAAUUGUUGCCGAUUUUGAUAAAUGCAAUGAGUGAGUUGUUGCUGUUGA---UGUU ........(((((((((...((((.((.(((..((......(((((((((.....))))))))).((((....))))))..)))))..))))))))))))).---.... ( -30.80) >DroPer_CAF1 4212 100 - 1 AGCCA------UAUAAACAACAAUAACAACAAGGCAGAGAGAGCAACAAUUUGUAAUUGCUGCGGAUUUUGAUAAAUGCAAUAUGUUUGCUGUUGCUGU---AGCCUUU .....------...................(((((....(.((((((((((((((.....)))))))).........((((.....)))).)))))).)---.))))). ( -21.50) >consensus AGCCA______CAACAAAAGCAACAACAACAAGGCAGACAGGGCAACAAUUUGCAAUUGUUGCUGAUUUUGAUAAAUGCAAUGAGUAUGCUGUUGCUGUUGUACCCGUU ....................(((((((.......((((...(((((((((.....)))))))))...))))....((((.....))))))))))).............. (-15.03 = -16.17 + 1.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:17 2006