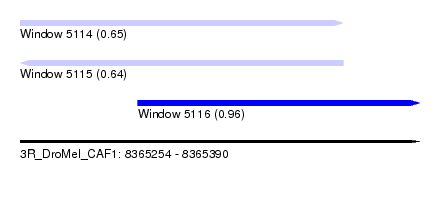
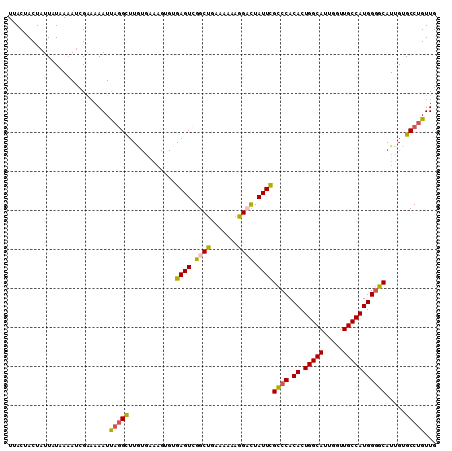
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,365,254 – 8,365,390 |
| Length | 136 |
| Max. P | 0.962049 |

| Location | 8,365,254 – 8,365,364 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 84.24 |
| Mean single sequence MFE | -30.45 |
| Consensus MFE | -24.45 |
| Energy contribution | -24.32 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8365254 110 + 27905053 UUACUACUAUUAUAAAAUCGAAAAAUCAGGCUUAUGAAAGUGUGAGUCGGCCGAAAAAAGGCUUAUUCGCCCACACUGGCAUUGGUUGCCAUGGGGCAUUGUGCCUGUUG ..........................(((((............((((.((((.......)))).))))((((.((.(((((.....))))))))))).....)))))... ( -35.00) >DroSec_CAF1 27453 110 + 1 UGACUACUAUUAUAAAAUGGGAAAAUUAGGCUUGUGAAAGUGUGAGUCGGCUGAAAAAAGGACUAUUCGCCCACACUGGCAAUGGUUGCCAUGGGGCAUUGUGCCUGUUG .(((..(((((....))))).....(((.((((....)))).))))))(((.(((..........)))((((.((.(((((.....))))))))))).....)))..... ( -29.60) >DroSim_CAF1 26137 100 + 1 UUACUACUAUUAUAAAAUCGAAAAAUUAGGCUU----------GAGUUGGCUGAAAAAAGGACUAUUUGCCCACACUGGCAUUGGUUGCCAUGGGGCAUUGUGCCUGUUG ..........................(((((..----------.((((..((......))))))...(((((.((.(((((.....))))))))))))....)))))... ( -25.40) >DroYak_CAF1 27962 109 + 1 UUACAACUAUUAUAAUAACGG-AAAUUACGUUUGUGGAAGAGAGAGUCGGCUCAAAAAAGGCUUAUUCGUCCACACUGGCAUUGGUUGCCAUGGCACAUUGUGCCCGUUG ...((((.........((((.-......))))((((((.(((.(((((...........))))).))).)))))).(((((.....))))).(((((...))))).)))) ( -31.80) >consensus UUACUACUAUUAUAAAAUCGAAAAAUUAGGCUUGUGAAAGUGUGAGUCGGCUGAAAAAAGGACUAUUCGCCCACACUGGCAUUGGUUGCCAUGGGGCAUUGUGCCUGUUG ..........................(((((............((((.((((.......)))).))))((((.((.(((((.....))))))))))).....)))))... (-24.45 = -24.32 + -0.13)

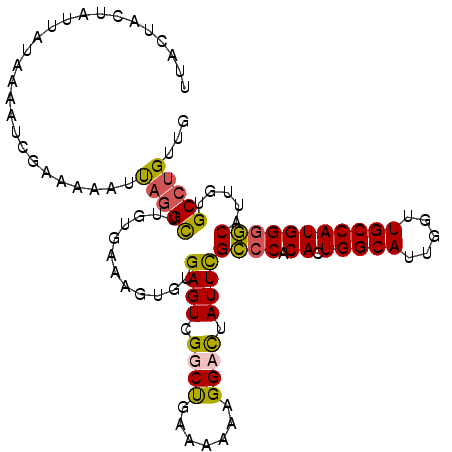
| Location | 8,365,254 – 8,365,364 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 84.24 |
| Mean single sequence MFE | -25.27 |
| Consensus MFE | -17.86 |
| Energy contribution | -17.55 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8365254 110 - 27905053 CAACAGGCACAAUGCCCCAUGGCAACCAAUGCCAGUGUGGGCGAAUAAGCCUUUUUUCGGCCGACUCACACUUUCAUAAGCCUGAUUUUUCGAUUUUAUAAUAGUAGUAA ...(((((....((((....))))....(((..((((((((((.....(((.......))))).))))))))..)))..))))).......................... ( -30.70) >DroSec_CAF1 27453 110 - 1 CAACAGGCACAAUGCCCCAUGGCAACCAUUGCCAGUGUGGGCGAAUAGUCCUUUUUUCAGCCGACUCACACUUUCACAAGCCUAAUUUUCCCAUUUUAUAAUAGUAGUCA .....(((.....(((((((((((.....))))).)).))))(((..........))).)))((((.((..................................)))))). ( -24.08) >DroSim_CAF1 26137 100 - 1 CAACAGGCACAAUGCCCCAUGGCAACCAAUGCCAGUGUGGGCAAAUAGUCCUUUUUUCAGCCAACUC----------AAGCCUAAUUUUUCGAUUUUAUAAUAGUAGUAA ....((((....((((((((((((.....))))).)).))))).......((......)).......----------..))))........................... ( -21.30) >DroYak_CAF1 27962 109 - 1 CAACGGGCACAAUGUGCCAUGGCAACCAAUGCCAGUGUGGACGAAUAAGCCUUUUUUGAGCCGACUCUCUCUUCCACAAACGUAAUUU-CCGUUAUUAUAAUAGUUGUAA ((((.(((((...))))).(((((.....))))).((((((.((....(.(((....))).).......)).))))))((((......-.)))).........))))... ( -25.00) >consensus CAACAGGCACAAUGCCCCAUGGCAACCAAUGCCAGUGUGGGCGAAUAAGCCUUUUUUCAGCCGACUCACACUUUCACAAGCCUAAUUUUCCGAUUUUAUAAUAGUAGUAA .....(((.....(((((((((((.....))))).)).))))(((........)))...)))................................................ (-17.86 = -17.55 + -0.31)

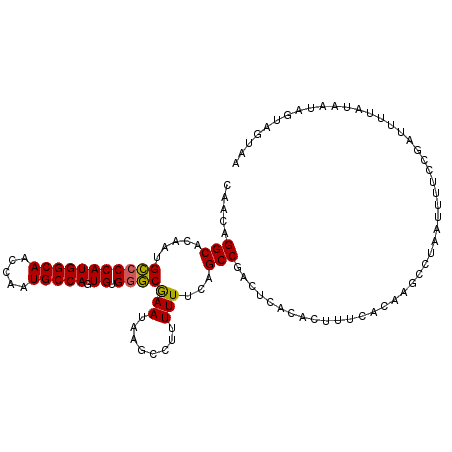
| Location | 8,365,294 – 8,365,390 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 74.10 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -15.48 |
| Energy contribution | -16.96 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.962049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
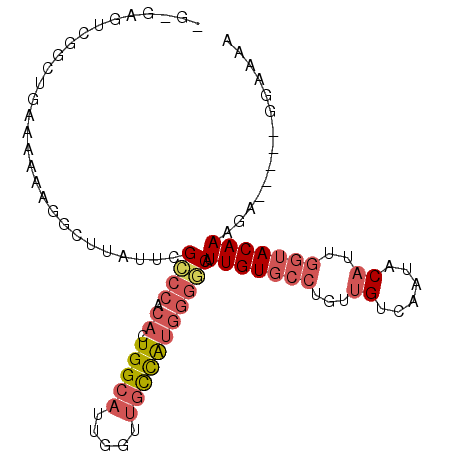
>3R_DroMel_CAF1 8365294 96 + 27905053 UGUGAGUCGGCCGAAAAAAGGCUUAUUCGCCCACACUGGCAUUGGUUGCCAUGGGGCAUUGUGCCUGUUGUCAAUACAUUGGUACAAAGA-----GGAAAA ...((((.((((.......)))).))))((((.((.(((((.....))))))))))).(((((((...(((....)))..)))))))...-----...... ( -36.50) >DroSec_CAF1 27493 96 + 1 UGUGAGUCGGCUGAAAAAAGGACUAUUCGCCCACACUGGCAAUGGUUGCCAUGGGGCAUUGUGCCUGUUGUCAAUACAUUGGUACAAAGG-----GUAAAA ....((((..((......))))))((((((((.((.(((((.....))))))))))).(((((((...(((....)))..))))))).))-----)).... ( -29.90) >DroSim_CAF1 26170 93 + 1 ---GAGUUGGCUGAAAAAAGGACUAUUUGCCCACACUGGCAUUGGUUGCCAUGGGGCAUUGUGCCUGUUGUCAAUACAUUGGUACAAAGG-----GUGAAA ---...((.(((...............(((((.((.(((((.....))))))))))))(((((((...(((....)))..)))))))..)-----)).)). ( -29.80) >DroYak_CAF1 28001 96 + 1 AGAGAGUCGGCUCAAAAAAGGCUUAUUCGUCCACACUGGCAUUGGUUGCCAUGGCACAUUGUGCCCGUUGUCAUUACAUUGGUACAAAGA-----UGAAAA ...((((.((((.......)))).))))(((((...(((((.....)))))))).)).(((((((...(((....)))..)))))))...-----...... ( -25.90) >DroAna_CAF1 28643 84 + 1 ----------------AAUUGCUGAUUUGUCAGAACUGGUAUUGGUGCUUGGGGGCCAUUGUUC-UCCAGCCAAUACAUUGCUACAAAGACACAAAGAAAA ----------------..(((.((.(((((.((.....((((((((....((((((....))))-))..))))))))....)))))))..)))))...... ( -21.40) >consensus _G_GAGUCGGCUGAAAAAAGGCUUAUUCGCCCACACUGGCAUUGGUUGCCAUGGGGCAUUGUGCCUGUUGUCAAUACAUUGGUACAAAGA_____GGAAAA ............................((((.((.(((((.....))))))))))).(((((((...((......))..))))))).............. (-15.48 = -16.96 + 1.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:28 2006