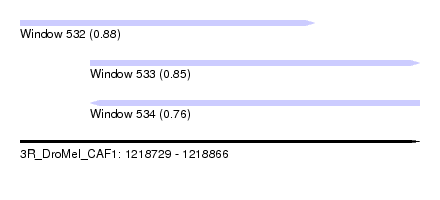
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,218,729 – 1,218,866 |
| Length | 137 |
| Max. P | 0.879649 |

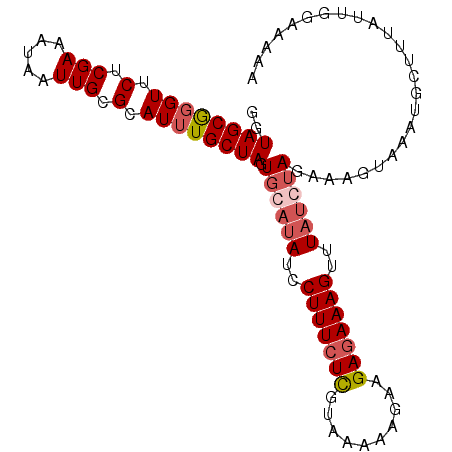
| Location | 1,218,729 – 1,218,830 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 79.04 |
| Mean single sequence MFE | -22.27 |
| Consensus MFE | -15.95 |
| Energy contribution | -17.33 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1218729 101 + 27905053 GGUAGCGGGUUCUCGAAAUAAUUGCGCAUUUGCUAGUGAAUAUCCUUUCUCUUAAACAGAAGAGAAAGUUUUCCUAGAAAUUAAAUGCUUUAUUGCAAAAU .(((((((.............))))(((((((((((.(((....(((((((((......)))))))))..)))))))....))))))).....)))..... ( -23.52) >DroSec_CAF1 9581 101 + 1 GGUAGCGGGUUCUCGAAAUAAUUGCGCAUUUGCUAGUGCAUAUCCUUUCUCGUAAAAAGAAGAGAAAGUUUAUGUAGAAAGUAAAUGCUUUAUUGGAAAAA ....((((.............))))(((((((((..((((((..(((((((..........)))))))..))))))...)))))))))............. ( -27.82) >DroSim_CAF1 9713 101 + 1 GGUAGCGGGUUCUCGAAAUAAUUGCGCAUUUGCUAGUGCAUAUCCUUUCUCGUAAAAAGAAGAGAAAGUUUAUGUAGAAAGUAAAUGCUUUAUUGGAAAAA ....((((.............))))(((((((((..((((((..(((((((..........)))))))..))))))...)))))))))............. ( -27.82) >DroYak_CAF1 13189 77 + 1 GGUAGCAUGUCCUCGAAAUAAUUGCGCAUUUGCUAUUCCAUAUUCUUUGUUGUUAAAAGAAGAGAAAGUUUAUCUAC------------------------ (((((((.((.(.(((.....))).).)).)))))..))...((((((.......))))))................------------------------ ( -9.90) >consensus GGUAGCGGGUUCUCGAAAUAAUUGCGCAUUUGCUAGUGCAUAUCCUUUCUCGUAAAAAGAAGAGAAAGUUUAUCUAGAAAGUAAAUGCUUUAUUGGAAAAA ..((((((((.(.(((.....))).).)))))))).((((((..(((((((..........)))))))..))))))......................... (-15.95 = -17.33 + 1.38)

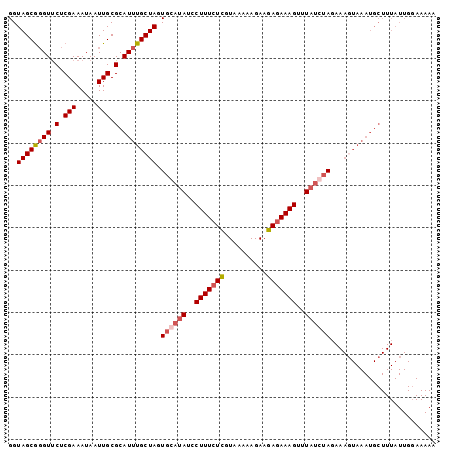
| Location | 1,218,753 – 1,218,866 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 90.72 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -21.33 |
| Energy contribution | -23.33 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.79 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1218753 113 + 27905053 CGCAUUUGCUAGUGAAUAUCCUUUCUCUUAAACAGAAGAGAAAGUUUUCCUAGAAAUUAAAUGCUUUAUUGCAAAAU-CUCAAACGGAAAAUGGA-UAUUUAUAAAUUUGAUUUA .((....))..((((((((((((((((((......))))))))(((((((...........(((......)))....-.......))))))))))-)))))))............ ( -27.65) >DroSec_CAF1 9605 115 + 1 CGCAUUUGCUAGUGCAUAUCCUUUCUCGUAAAAAGAAGAGAAAGUUUAUGUAGAAAGUAAAUGCUUUAUUGGAAAAACAUCAAACGGAAAAUGGAAUUUUUUUAAAUUUGAGUUA .(((((((((..((((((..(((((((..........)))))))..))))))...)))))))))...........(((.(((((..(((((.......)))))...)))))))). ( -31.30) >DroSim_CAF1 9737 115 + 1 CGCAUUUGCUAGUGCAUAUCCUUUCUCGUAAAAAGAAGAGAAAGUUUAUGUAGAAAGUAAAUGCUUUAUUGGAAAAACAUGAAACGGAAAAUGGAAUUUUUUUAAAUUUGAGUUA .(((((((((..((((((..(((((((..........)))))))..))))))...)))))))))....((((((((((((..........)))...))))))))).......... ( -28.80) >consensus CGCAUUUGCUAGUGCAUAUCCUUUCUCGUAAAAAGAAGAGAAAGUUUAUGUAGAAAGUAAAUGCUUUAUUGGAAAAACAUCAAACGGAAAAUGGAAUUUUUUUAAAUUUGAGUUA .(((((((((..((((((..(((((((..........)))))))..))))))...)))))))))...............(((((..(((((.......)))))...))))).... (-21.33 = -23.33 + 2.00)

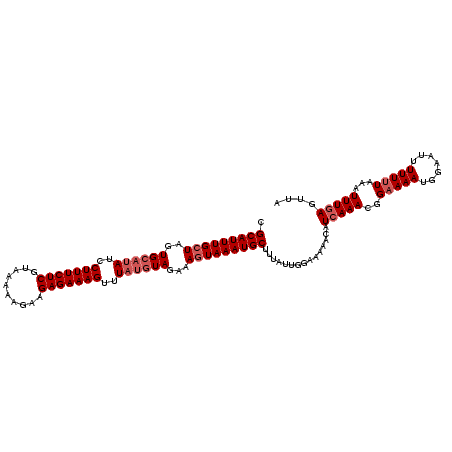
| Location | 1,218,753 – 1,218,866 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 90.72 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -16.30 |
| Energy contribution | -16.97 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1218753 113 - 27905053 UAAAUCAAAUUUAUAAAUA-UCCAUUUUCCGUUUGAG-AUUUUGCAAUAAAGCAUUUAAUUUCUAGGAAAACUUUCUCUUCUGUUUAAGAGAAAGGAUAUUCACUAGCAAAUGCG ...............((((-(((.((((((....(((-(((.(((......)))...))))))..)))))).((((((((......))))))))))))))).....((....)). ( -28.20) >DroSec_CAF1 9605 115 - 1 UAACUCAAAUUUAAAAAAAUUCCAUUUUCCGUUUGAUGUUUUUCCAAUAAAGCAUUUACUUUCUACAUAAACUUUCUCUUCUUUUUACGAGAAAGGAUAUGCACUAGCAAAUGCG .(((((((((..((((........))))..)))))).)))...........((((((.((.....((((..(((((((..........)))))))..))))....)).)))))). ( -22.30) >DroSim_CAF1 9737 115 - 1 UAACUCAAAUUUAAAAAAAUUCCAUUUUCCGUUUCAUGUUUUUCCAAUAAAGCAUUUACUUUCUACAUAAACUUUCUCUUCUUUUUACGAGAAAGGAUAUGCACUAGCAAAUGCG ...................................................((((((.((.....((((..(((((((..........)))))))..))))....)).)))))). ( -18.50) >consensus UAACUCAAAUUUAAAAAAAUUCCAUUUUCCGUUUGAUGUUUUUCCAAUAAAGCAUUUACUUUCUACAUAAACUUUCUCUUCUUUUUACGAGAAAGGAUAUGCACUAGCAAAUGCG ....((((((..((((........))))..))))))...............((((((.((.....((((..(((((((..........)))))))..))))....)).)))))). (-16.30 = -16.97 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:15 2006