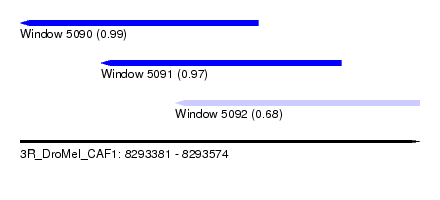
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,293,381 – 8,293,574 |
| Length | 193 |
| Max. P | 0.991556 |

| Location | 8,293,381 – 8,293,496 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.99 |
| Mean single sequence MFE | -24.05 |
| Consensus MFE | -15.30 |
| Energy contribution | -15.67 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8293381 115 - 27905053 AAGUGAACACGUCGCUAAGCGAAAGCUAAGCAAAUAAACAAGCGCAGCUGAACAAGCUAAA----CAAUCUGCAGUAAAGU-GCAAGUUAAAGUGAAUCAAUUAAAAGUAACCAGCAACC ...(((.(((...(((.(((....))).))).....(((..((((((((.....))))..(----(........))...))-))..)))...)))..))).................... ( -23.70) >DroSim_CAF1 5013 117 - 1 AAGUGAACACAUCGCUAAGCGAAAGCUAAGCAAACAAACAAGCGCAGCUGAACAAGCUAAACAAACAAUCUGCAUUGAAGU-GCAAGUUAAAGUGAAUCAAUUU-AAGUAACCAGCA-CC ...(((.(((...(((.(((....))).))).....(((..((((((((.....)))).......((((....))))..))-))..)))...)))..)))....-............-.. ( -25.30) >DroEre_CAF1 5015 100 - 1 AAGUGAACACAUCGCUGAGCGAAAGCUAAGCAACCAAAUAAGCGCAGCUGAUCAAGCUAAA----CAAACUGCAGUGAAGU-------------G---CAAGUAAAAUAAACUAGCAAUC .......(((...(((.(((....))).)))..............((((.....))))...----.........)))...(-------------(---(.(((.......))).)))... ( -21.80) >DroYak_CAF1 4950 116 - 1 AACUGAAAACAUCGCUAAGCGAAAGCUAAGCAAACAAACAAGCGCAGCUGAACAAGCUAAA----CAAUCUGCAGUAACGUUGCAAGUUAAAGUGAAUCAAGUAACAGUAAUUAGGAAAC .((((........(((.(((....))).)))...........(((((((.....))))...----.((..(((((.....)))))..))...)))..........))))........... ( -25.40) >consensus AAGUGAACACAUCGCUAAGCGAAAGCUAAGCAAACAAACAAGCGCAGCUGAACAAGCUAAA____CAAUCUGCAGUAAAGU_GCAAGUUAAAGUGAAUCAAGUAAAAGUAACCAGCAACC .((((........(((.(((....))).)))..............((((.....))))..............))))............................................ (-15.30 = -15.67 + 0.37)

| Location | 8,293,420 – 8,293,536 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.72 |
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -15.50 |
| Energy contribution | -17.75 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8293420 116 - 27905053 GCGCUUCGUCUACGGAGCGACAAUUCAAUUUAAACAAGCAAAGUGAACACGUCGCUAAGCGAAAGCUAAGCAAAUAAACAAGCGCAGCUGAACAAGCUAAA----CAAUCUGCAGUAAAG ((((((((....))))))(((..((((.(((........))).))))...)))(((.(((....))).)))............))((((.....))))...----............... ( -29.70) >DroPse_CAF1 5303 102 - 1 CCGCCAGUUUCUCUUCGCAGCAAUUCAAUUCAAACAAGCGAAGAGAACACAUCUCAAAAGUGAAACAAAGCGAGCUACCAA------------AAGCCAAA----ACAACUGCAAAAA-- ..((.(((((((((((((...................))))))))).(((.........))).........(.(((.....------------.))))...----..)))))).....-- ( -19.81) >DroSec_CAF1 4983 120 - 1 CCGCUUCGUUCCCAGAGCGUCAAUUCAAUUCAAACAAGCAAAGUGAACACAUCGCUAAGCGAAAGCUAAGCAAACAAACAAGCGCAGCUGAACAAGCUAAACAAACAAUCUGCAUUAAAG .(((((........)))))..................(((...((........(((.(((....))).)))..............((((.....)))).......))...)))....... ( -23.70) >DroSim_CAF1 5050 120 - 1 CCGCUUCGUUCCCAGAGCGUCAAUUCAAUUCAAACAAGCAAAGUGAACACAUCGCUAAGCGAAAGCUAAGCAAACAAACAAGCGCAGCUGAACAAGCUAAACAAACAAUCUGCAUUGAAG ...(((((....((((((((......................(....).....(((.(((....))).)))..........))))((((.....))))..........))))...))))) ( -26.90) >DroEre_CAF1 5039 116 - 1 CUGCUUGUUUCUUUUAGCGUCAUUUCAAUUCAAAGAAGCAAAGUGAACACAUCGCUGAGCGAAAGCUAAGCAACCAAAUAAGCGCAGCUGAUCAAGCUAAA----CAAACUGCAGUGAAG ((((..((((..((((((.((((((...(((...)))...))))))....((((((((((....)))..((..........)).)))).)))...))))))----.)))).))))..... ( -28.70) >DroYak_CAF1 4990 116 - 1 CCGCUUGGUUCUCUUAGCGUCAAUUCAAUUCAAACAAGCAAACUGAAAACAUCGCUAAGCGAAAGCUAAGCAAACAAACAAGCGCAGCUGAACAAGCUAAA----CAAUCUGCAGUAACG .(((((((((.((...((...................)).....)).)))...(((.(((....))).))).......)))))).((((.....))))...----............... ( -24.91) >consensus CCGCUUCGUUCUCUGAGCGUCAAUUCAAUUCAAACAAGCAAAGUGAACACAUCGCUAAGCGAAAGCUAAGCAAACAAACAAGCGCAGCUGAACAAGCUAAA____CAAUCUGCAGUAAAG .(((((((....)))))))..................(((.............(((.(((....))).)))..............((((.....))))............)))....... (-15.50 = -17.75 + 2.25)

| Location | 8,293,456 – 8,293,574 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.85 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -16.49 |
| Energy contribution | -18.13 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
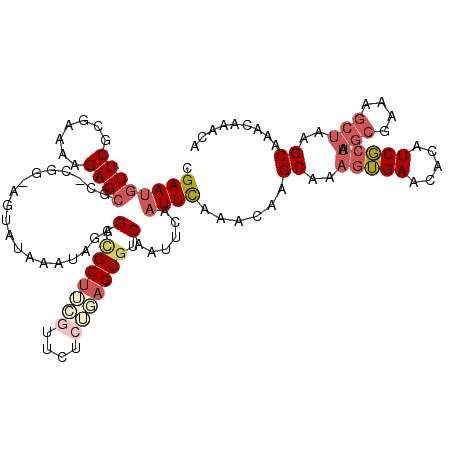
>3R_DroMel_CAF1 8293456 118 - 27905053 CGAAUGUUCGCGAAAAGAGCGC-AGC-AGUAUAAAUAGAGGCGCUUCGUCUACGGAGCGACAAUUCAAUUUAAACAAGCAAAGUGAACACGUCGCUAAGCGAAAGCUAAGCAAAUAAACA .((.((((((((.......)))-.((-.((.(((((.(((.(((((((....)))))))....))).))))).))..)).....)))))..))(((.(((....))).)))......... ( -37.20) >DroSec_CAF1 5023 118 - 1 CGAAUGUUCGCGAAAAGAGCGC-CGG-AGUAUAAAUAGAGCCGCUUCGUUCCCAGAGCGUCAAUUCAAUUCAAACAAGCAAAGUGAACACAUCGCUAAGCGAAAGCUAAGCAAACAAACA .((.((((((((((..(((((.-(..-............).)))))..))).....((...................))...)))))))..))(((.(((....))).)))......... ( -28.25) >DroSim_CAF1 5090 119 - 1 CGAAUGUUCGCGAAAAGAGCGCCCGG-AGUAUAAAUAGAGCCGCUUCGUUCCCAGAGCGUCAAUUCAAUUCAAACAAGCAAAGUGAACACAUCGCUAAGCGAAAGCUAAGCAAACAAACA .((.(((((((.....(((((..(((-..(.......)..)))...))))).....((...................))...)))))))..))(((.(((....))).)))......... ( -28.21) >DroEre_CAF1 5075 119 - 1 CGAAUGUUCUCGAAAGGAGCGC-CGGGUGUAUAAAUAGCGCUGCUUGUUUCUUUUAGCGUCAUUUCAAUUCAAAGAAGCAAAGUGAACACAUCGCUGAGCGAAAGCUAAGCAACCAAAUA .((.(((((.(..(((.(((((-...((......)).))))).)))((((((((.................))))))))...).)))))..))(((.(((....))).)))......... ( -32.83) >DroYak_CAF1 5026 118 - 1 CGAAUGUUCGCGAAAAGAGCGC-CGG-AGUAUAAAUAGGGCCGCUUGGUUCUCUUAGCGUCAAUUCAAUUCAAACAAGCAAACUGAAAACAUCGCUAAGCGAAAGCUAAGCAAACAAACA .((((...(((...(((((.((-((.-(((............))))))).))))).)))...))))...........................(((.(((....))).)))......... ( -29.20) >DroPer_CAF1 5479 118 - 1 CGAAGUUUCGAACGGAGAGCGG-CCA-GGUAUAAAUACAGCCGCCAGUUUCUCUUCGCAGCAAUUCAAUUCAAACAAGCGAAGAGAACACAUCUCAAAAGUGAAACAAAGCGAGCUACCA ....((((((....((((((((-(..-.(((....))).)))))..((((((((((((...................)))))))))).)).)))).....)))))).............. ( -35.81) >consensus CGAAUGUUCGCGAAAAGAGCGC_CGG_AGUAUAAAUAGAGCCGCUUCGUUCUCUGAGCGUCAAUUCAAUUCAAACAAGCAAAGUGAACACAUCGCUAAGCGAAAGCUAAGCAAACAAACA .((((((((.......))))...................(.(((((((....))))))).)......))))......((..(((((.....))))).(((....)))..))......... (-16.49 = -18.13 + 1.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:05 2006