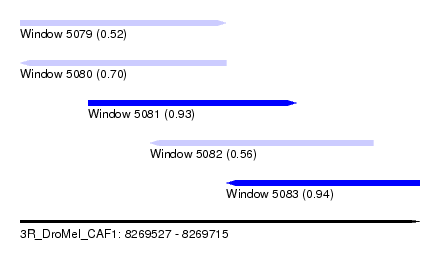
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,269,527 – 8,269,715 |
| Length | 188 |
| Max. P | 0.939726 |

| Location | 8,269,527 – 8,269,624 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -16.70 |
| Consensus MFE | -10.18 |
| Energy contribution | -10.32 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
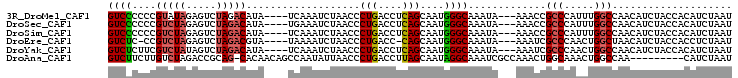
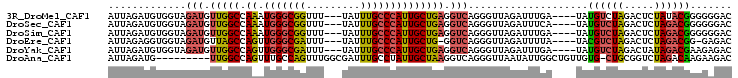
>3R_DroMel_CAF1 8269527 97 + 27905053 GUCCCCCCGUAUAGAGUCUAGACAUA----UCAAAUCUAACCCUGACCUCAGCAAUGGGCAAAUA---AAACCGCCCAUUUGGCCAACAUCUACCACAUCUAAU ...........((((((.((((....----............(((....))).(((((((.....---.....))))))).........))))..)).)))).. ( -14.80) >DroSec_CAF1 9261 97 + 1 GUCCCCCCGUCUAGAGUCUAGACAUA----UGAAAUCUAACCCUGACCUCAGCAAUGGGCAAAUA---AAACCGCCCAUUUGGCCAACAUCUACCACAUCUAAU ........((((((...))))))...----(((..((.......))..)))((.((((((.....---.....))))))...)).................... ( -17.90) >DroSim_CAF1 9619 97 + 1 GUCCCCCCGUCUAGAGUCUAGACAUA----UCAAAUCUAACCCUGACCUCAGCAAUGGGCAAAUA---AAACCGCCCAUUUGGCCAACAUCUACCACAUCUAAU ......((((((((...))))))...----............(((....))).(((((((.....---.....))))))).))..................... ( -17.50) >DroEre_CAF1 7904 95 + 1 GUCUC-CCGUCUAGAGUCUAGACGUA----UAAAAUCUAACCCUGACC-CAGCAAUGGGCAAAUA---AAAUCGCCCAACUGGCUAACAUCUACCACCUCUAAU .....-.(((((((...)))))))..----.................(-(((...(((((.....---.....))))).))))..................... ( -19.60) >DroYak_CAF1 9164 97 + 1 GUCUCUUCGUCUAUAGUCUAGACAUA----UCAAAUCUAACCCUGACCUCAGCAAUGGGCAAAUA---AAAUCGCCCAACUGGCCAACAUCUACCACAUCUAAU (((.....(((((.....)))))...----............(((....)))...(((((.....---.....)))))...))).................... ( -14.80) >DroAna_CAF1 11247 94 + 1 GUCUUCUUGUCUAGACCGCAG-CACAACAGCCAAUAUUAACCCUGACCUUAGCAAUAGGCAAAUCGCCAAACUGGCAAACUGGCCAA---------CAUCUAAU ((((........)))).....-.......((((..........((.(((.......)))))....(((.....)))....))))...---------........ ( -15.60) >consensus GUCCCCCCGUCUAGAGUCUAGACAUA____UCAAAUCUAACCCUGACCUCAGCAAUGGGCAAAUA___AAACCGCCCAACUGGCCAACAUCUACCACAUCUAAU ((((....(((((.....)))))...................(((....)))....)))).............(((.....))).................... (-10.18 = -10.32 + 0.14)
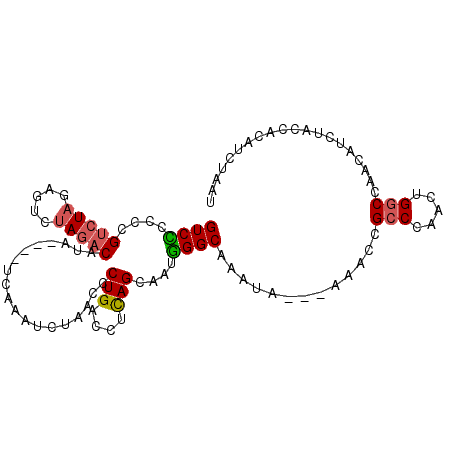
| Location | 8,269,527 – 8,269,624 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -18.89 |
| Energy contribution | -18.92 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8269527 97 - 27905053 AUUAGAUGUGGUAGAUGUUGGCCAAAUGGGCGGUUU---UAUUUGCCCAUUGCUGAGGUCAGGGUUAGAUUUGA----UAUGUCUAGACUCUAUACGGGGGGAC .(((((((((.(((((.((((((.(((((((((...---...))))))))).(((....)))))))))))))).----))))))))).((((.....))))... ( -32.20) >DroSec_CAF1 9261 97 - 1 AUUAGAUGUGGUAGAUGUUGGCCAAAUGGGCGGUUU---UAUUUGCCCAUUGCUGAGGUCAGGGUUAGAUUUCA----UAUGUCUAGACUCUAGACGGGGGGAC .(((((((((..((((.((((((.(((((((((...---...))))))))).(((....)))))))))))))..----))))))))).((((.....))))... ( -30.70) >DroSim_CAF1 9619 97 - 1 AUUAGAUGUGGUAGAUGUUGGCCAAAUGGGCGGUUU---UAUUUGCCCAUUGCUGAGGUCAGGGUUAGAUUUGA----UAUGUCUAGACUCUAGACGGGGGGAC .(((((((((.(((((.((((((.(((((((((...---...))))))))).(((....)))))))))))))).----))))))))).((((.....))))... ( -32.20) >DroEre_CAF1 7904 95 - 1 AUUAGAGGUGGUAGAUGUUAGCCAGUUGGGCGAUUU---UAUUUGCCCAUUGCUG-GGUCAGGGUUAGAUUUUA----UACGUCUAGACUCUAGACGG-GAGAC .((((((....(((((((...((((((((((((...---...)))))))..))))-)(((.......)))....----.)))))))..))))))....-..... ( -30.10) >DroYak_CAF1 9164 97 - 1 AUUAGAUGUGGUAGAUGUUGGCCAGUUGGGCGAUUU---UAUUUGCCCAUUGCUGAGGUCAGGGUUAGAUUUGA----UAUGUCUAGACUAUAGACGAAGAGAC (((((((.(..(.(((.(..((....(((((((...---...)))))))..))..).)))...)..).))))))----).((((((.....))))))....... ( -30.00) >DroAna_CAF1 11247 94 - 1 AUUAGAUG---------UUGGCCAGUUUGCCAGUUUGGCGAUUUGCCUAUUGCUAAGGUCAGGGUUAAUAUUGGCUGUUGUG-CUGCGGUCUAGACAAGAAGAC ....((((---------((((((.....(((...((((((((......)))))))))))...))))))))))((((((....-..))))))............. ( -25.90) >consensus AUUAGAUGUGGUAGAUGUUGGCCAAAUGGGCGGUUU___UAUUUGCCCAUUGCUGAGGUCAGGGUUAGAUUUGA____UAUGUCUAGACUCUAGACGGGGAGAC .............(((.(((((.((.(((((((.........)))))))))))))).)))....................((((((.....))))))....... (-18.89 = -18.92 + 0.03)
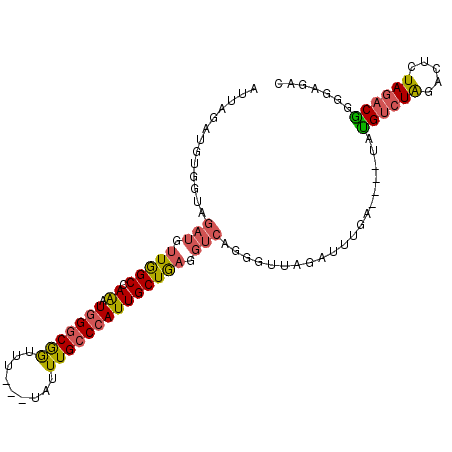
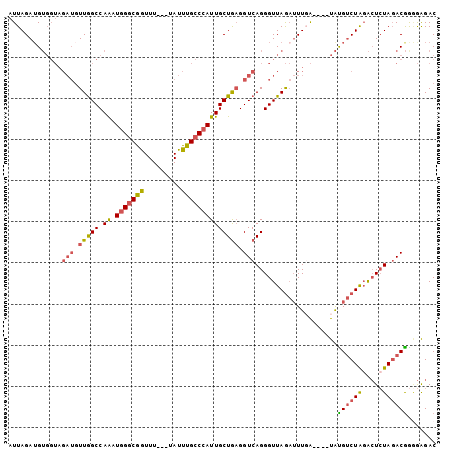
| Location | 8,269,559 – 8,269,657 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 97.28 |
| Mean single sequence MFE | -14.00 |
| Consensus MFE | -14.00 |
| Energy contribution | -14.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8269559 98 + 27905053 CUAACCCUGACCUCAGCAAUGGGCAAAUAAAACCGCCCAUUUGGCCAACAUCUACCACAUCUAAUCUGCUAAUGAGAAUACACGCACAUACCACACAU ...........(((((((((((((..........)))))))(((..........)))..........))...))))...................... ( -14.00) >DroSec_CAF1 9293 98 + 1 CUAACCCUGACCUCAGCAAUGGGCAAAUAAAACCGCCCAUUUGGCCAACAUCUACCACAUCUAAUCUGCUAAUGAGAAUACGCACACACACCACACAU ...........(((((((((((((..........)))))))(((..........)))..........))...))))...................... ( -14.00) >DroSim_CAF1 9651 98 + 1 CUAACCCUGACCUCAGCAAUGGGCAAAUAAAACCGCCCAUUUGGCCAACAUCUACCACAUCUAAUCUGCUAAUGAGAAUACGCACACACACCACCCAU ...........(((((((((((((..........)))))))(((..........)))..........))...))))...................... ( -14.00) >consensus CUAACCCUGACCUCAGCAAUGGGCAAAUAAAACCGCCCAUUUGGCCAACAUCUACCACAUCUAAUCUGCUAAUGAGAAUACGCACACACACCACACAU ...........(((((((((((((..........)))))))(((..........)))..........))...))))...................... (-14.00 = -14.00 + 0.00)


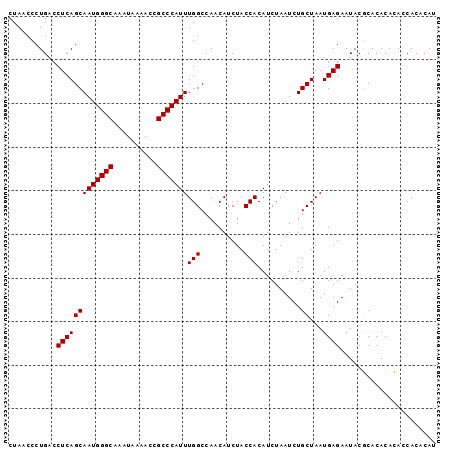
| Location | 8,269,588 – 8,269,693 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 93.02 |
| Mean single sequence MFE | -32.93 |
| Consensus MFE | -27.51 |
| Energy contribution | -29.07 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8269588 105 - 27905053 UUGCAGGUGUGUGCGUGCGCGCGCCUAUACAUACAUAUGUGUGGUAUGUGCGUGUAUUCUCAUUAGCAGAUUAGAUGUGGUAGAUGUUGGCCAAAUGGGCGGUUU ((((..(((.(...((((((((((((((((((....))))))))...)))))))))).).)))..))))....................(((.....)))..... ( -32.60) >DroSec_CAF1 9322 105 - 1 UUGCAGGUGUGUGCGUGCGCGCGCCAAUACAUACAUAUGUGUGGUGUGUGUGCGUAUUCUCAUUAGCAGAUUAGAUGUGGUAGAUGUUGGCCAAAUGGGCGGUUU ((((..(((.(((((..(((((((((.(((((....))))))))))))))..)))))...)))..))))....................(((.....)))..... ( -40.10) >DroSim_CAF1 9680 99 - 1 UUGCAGGUGUGUGCGUG------CCUAUACAUACAUAUGGGUGGUGUGUGUGCGUAUUCUCAUUAGCAGAUUAGAUGUGGUAGAUGUUGGCCAAAUGGGCGGUUU ((((..(((.(((((..------(.(((((.(((......)))))))).)..)))))...)))..))))....................(((.....)))..... ( -26.10) >consensus UUGCAGGUGUGUGCGUGCGCGCGCCUAUACAUACAUAUGUGUGGUGUGUGUGCGUAUUCUCAUUAGCAGAUUAGAUGUGGUAGAUGUUGGCCAAAUGGGCGGUUU ((((..(((.(((((((((((.(((.((((((....))))))))).)))))))))))...)))..))))....................(((.....)))..... (-27.51 = -29.07 + 1.56)

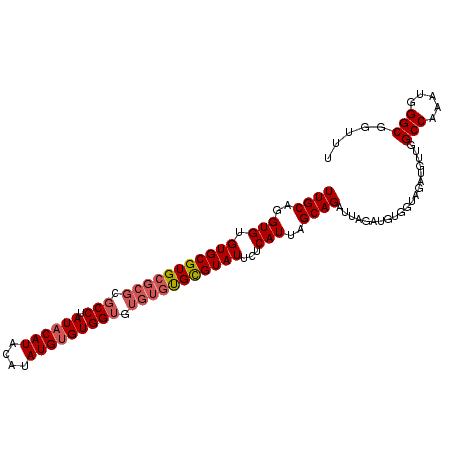
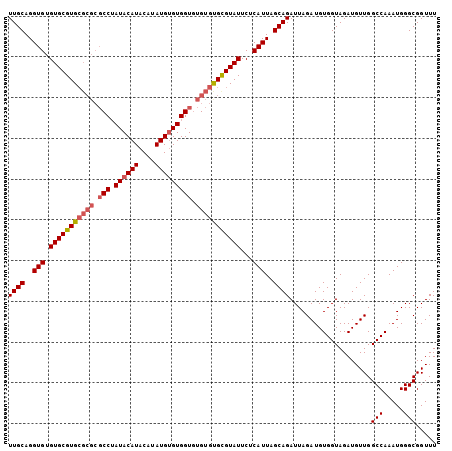
| Location | 8,269,624 – 8,269,715 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 91.21 |
| Mean single sequence MFE | -33.27 |
| Consensus MFE | -27.37 |
| Energy contribution | -28.93 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.01 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8269624 91 - 27905053 AGCUUUAGAUUAGUUAAAGCUUUUGCAGGUGUGUGCGUGCGCGCGCCUAUACAUACAUAUGUGUGGUAUGUGCGUGUAUUCUCAUUAGCAG ((((((((.....))))))))..(((..(((.(...((((((((((((((((((....))))))))...)))))))))).).)))..))). ( -33.60) >DroSec_CAF1 9358 91 - 1 AGCUUUACAUUAGUUAAAGCUUUUGCAGGUGUGUGCGUGCGCGCGCCAAUACAUACAUAUGUGUGGUGUGUGUGCGUAUUCUCAUUAGCAG (((((((.......)))))))..(((..(((.(((((..(((((((((.(((((....))))))))))))))..)))))...)))..))). ( -40.10) >DroSim_CAF1 9716 85 - 1 AGCUUUACAUUAGUUAAAGCUUUUGCAGGUGUGUGCGUG------CCUAUACAUACAUAUGGGUGGUGUGUGUGCGUAUUCUCAUUAGCAG (((((((.......)))))))..(((..(((.(((((..------(.(((((.(((......)))))))).)..)))))...)))..))). ( -26.10) >consensus AGCUUUACAUUAGUUAAAGCUUUUGCAGGUGUGUGCGUGCGCGCGCCUAUACAUACAUAUGUGUGGUGUGUGUGCGUAUUCUCAUUAGCAG (((((((.......)))))))..(((..(((.(((((((((((.(((.((((((....))))))))).)))))))))))...)))..))). (-27.37 = -28.93 + 1.56)

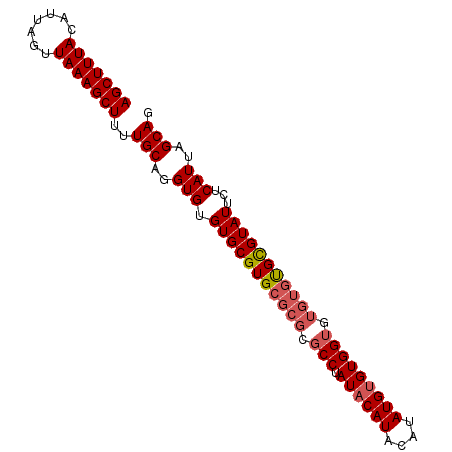
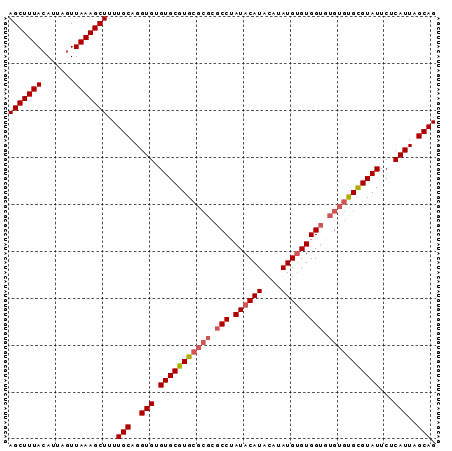
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:56 2006