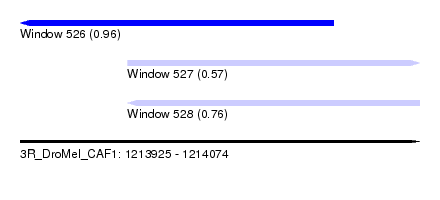
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,213,925 – 1,214,074 |
| Length | 149 |
| Max. P | 0.963946 |

| Location | 1,213,925 – 1,214,042 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.05 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -31.00 |
| Energy contribution | -31.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1213925 117 - 27905053 --AAUGCAUUCAGCCGUUA-AAUCGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCCUUUAGGCCACACGGUCCGGUUGAAACGUCCCGUUUGUCCC --......((((((((...-....(((((((...(((((((((.(((.......))).)))).)))))...((((........))))..)))))))))))))))................ ( -40.11) >DroVir_CAF1 4334 111 - 1 CAUAAAAAA--------AA-AAACGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCCAUUAGGCCACACGGUCCCGUUGUAAAGCGGCGUUUGUUGC .........--------..-(((((((((((...(((((((((.(((.......))).)))).)))))...((((........))))..)))))))(((((....))))).))))..... ( -38.20) >DroGri_CAF1 4747 109 - 1 --AAUACAA--------AU-CUUCGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCCAUUAGGCCACACGGUCCUGUUGUAAAGCGCCGUUAGUCGC --..(((((--------..-....(((((((...(((((((((.(((.......))).)))).)))))...((((........))))..)))))))...)))))..(((.(....).))) ( -35.70) >DroEre_CAF1 5695 105 - 1 ---------------CUUAUAAUCGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCCUUUAGGCCACACGGUCCCGUUGAAACGUCACGUUUGUCCC ---------------.........(((((((...(((((((((.(((.......))).)))).)))))...((((........))))..)))))))......(((((...)))))..... ( -32.30) >DroWil_CAF1 2733 109 - 1 --AAUAUAAUC---------AUUCGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCCUUUAGGCCACACGGUCCUUUUGCAUCGAACAGUUACUUUC --.........---------....(((((((...(((((((((.(((.......))).)))).)))))...((((........))))..)))))))........................ ( -31.00) >DroAna_CAF1 4702 113 - 1 --AGUAAAUU--GCA--GA-AGACGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCCUUUAGGCCACACGGUCCUGUUGAAACGUCACGUUUGUUCC --........--(((--((-.((((((((((...(((((((((.(((.......))).)))).)))))...((((........))))..)))))))..((....)))))...)))))... ( -35.60) >consensus __AAUAAAA________AA_AAUCGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCCUUUAGGCCACACGGUCCUGUUGAAACGUCCCGUUUGUCCC ........................(((((((...(((((((((.(((.......))).)))).)))))...((((........))))..)))))))........................ (-31.00 = -31.00 + 0.00)

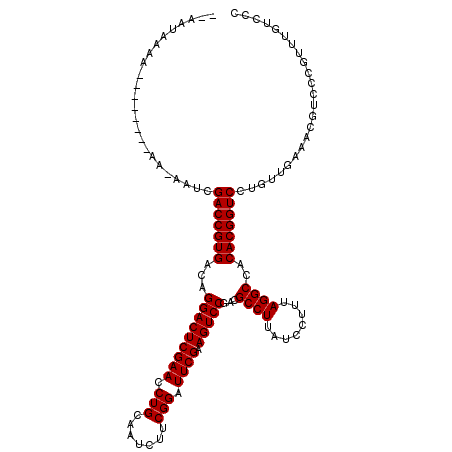
| Location | 1,213,965 – 1,214,074 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 74.43 |
| Mean single sequence MFE | -26.17 |
| Consensus MFE | -17.90 |
| Energy contribution | -17.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1213965 109 + 27905053 GGAUAAGGCGUCGGACUUCGAAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCGAUU-UAACGGCUGAAUGCAUU--UUUUGCACCGACUGGCUUGUUUAAACUUCUCU (((((((.((((((.(((((....)))))...(((((.......)))))..((((((...-...))))))..((((..--...))))))))).).))))))).......... ( -35.90) >DroVir_CAF1 4374 89 + 1 GGAUAAGGCGUCGGACUUCGAAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCGUUU-UU--------UUUUUUAUGUUUC--------------UUUUCAAUUUUUUU .(((..((((..(((((.((((((.(.......).)))))))))))))))...)))....-..--------.............--------------.............. ( -17.90) >DroGri_CAF1 4787 87 + 1 GGAUAAGGCGUCGGACUUCGAAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCGAAG-AU--------UUGUAUU--UUUU--------------UUUCUAAAUUGCAC .(((..((((..(((((.((((((.(.......).)))))))))))))))...)))....-..--------.((((.(--((..--------------.....))).)))). ( -19.20) >DroSim_CAF1 4980 109 + 1 GGAUAAGGCGUCGGACUUCGAAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCGAUU-UACCGGCUAAAUGCAUU--UUUUGCAUCAAGUCGGUUGUUUAAACUUCUUU .(((..((((..(((((.((((((.(.......).)))))))))))))))...)))(((.-.(((((((..(((((..--...)))))..))))))).)))........... ( -33.70) >DroYak_CAF1 5756 110 + 1 GGAUAAGGCGUCGGACUUCGAAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCGAUUAUAACUGCUAAGUGCAUU--UUUUGCAAUAAGUGGACCGUUCUAACUUUUUU .((((.(((.((((((((..((((....))))..))))))))))).))))((((((.(((...(((....)))(((..--...)))....))).))))))............ ( -31.90) >DroMoj_CAF1 4786 87 + 1 GGAUAAGGCGUCGGACUUCGAAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCGUAU-CA--------UACUUUU--UUUU--------------UUUUUAAUUUUCAG .((((.(((((.(((((.((((((.(.......).)))))))))))....)).))).)))-).--------.......--....--------------.............. ( -18.40) >consensus GGAUAAGGCGUCGGACUUCGAAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCGAUU_UA________UUGCAUU__UUUU______________GUUUUAACUUUUUU .(((..((((..(((((.((((((.(.......).)))))))))))))))...)))........................................................ (-17.90 = -17.90 + 0.00)

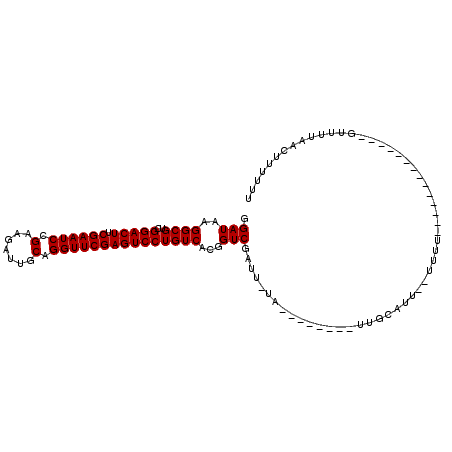
| Location | 1,213,965 – 1,214,074 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 74.43 |
| Mean single sequence MFE | -20.62 |
| Consensus MFE | -15.10 |
| Energy contribution | -15.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
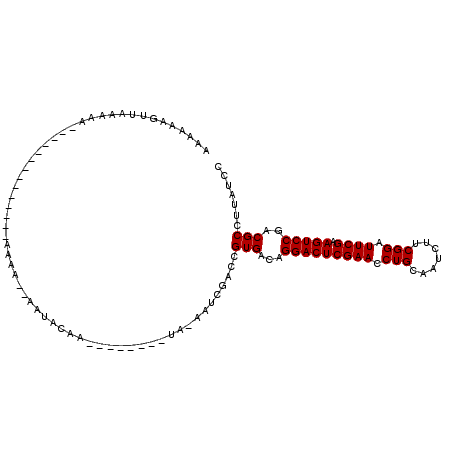
>3R_DroMel_CAF1 1213965 109 - 27905053 AGAGAAGUUUAAACAAGCCAGUCGGUGCAAAA--AAUGCAUUCAGCCGUUA-AAUCGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCC .(((..(((...........(((((((((...--..))))))..((.(((.-....))).))))).(((((((((.(((.......))).)))).))))))))..))).... ( -23.70) >DroVir_CAF1 4374 89 - 1 AAAAAAAUUGAAAA--------------GAAACAUAAAAAA--------AA-AAACGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCC ..............--------------.............--------..-........(((...(((((((((.(((.......))).)))).)))))..)))....... ( -15.10) >DroGri_CAF1 4787 87 - 1 GUGCAAUUUAGAAA--------------AAAA--AAUACAA--------AU-CUUCGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCC ..............--------------....--.......--------..-........(((...(((((((((.(((.......))).)))).)))))..)))....... ( -15.10) >DroSim_CAF1 4980 109 - 1 AAAGAAGUUUAAACAACCGACUUGAUGCAAAA--AAUGCAUUUAGCCGGUA-AAUCGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCC .(((...........((((.((.((((((...--..)))))).)).)))).-........(((...(((((((((.(((.......))).)))).)))))..)))))).... ( -28.50) >DroYak_CAF1 5756 110 - 1 AAAAAAGUUAGAACGGUCCACUUAUUGCAAAA--AAUGCACUUAGCAGUUAUAAUCGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCC ......(((...((((((.....(((((.((.--.......)).))))).......))))))....(((((((((.(((.......))).)))).))))))))......... ( -26.00) >DroMoj_CAF1 4786 87 - 1 CUGAAAAUUAAAAA--------------AAAA--AAAAGUA--------UG-AUACGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCC ..............--------------....--.......--------.(-(((.(..(((....(((((((((.(((.......))).)))).))))).))).).)))). ( -15.30) >consensus AAAAAAGUUAAAAA______________AAAA__AAUACAA________UA_AAUCGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUUCGAAGUCCGACGCCUUAUCC ............................................................(((...(((((((((.(((.......))).)))).)))))..)))....... (-15.10 = -15.10 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:09 2006