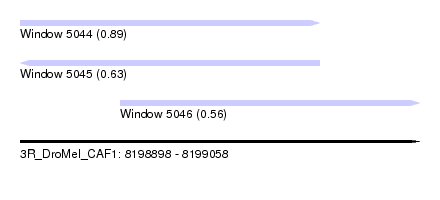
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,198,898 – 8,199,058 |
| Length | 160 |
| Max. P | 0.892402 |

| Location | 8,198,898 – 8,199,018 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -39.13 |
| Consensus MFE | -35.40 |
| Energy contribution | -34.90 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8198898 120 + 27905053 UACGCUGAAAGGGGAGCAAAUGAAUCCCGAUUUCAUCAAGAUCAACCCGCAGCACUCGAUUCCCACGCUGGUAGACAAUGGUUUCACCAUUUGGGAGUCGCGUGCCAUUCUCGUUUACCU ...((.....((((..(....)..))))(((((.....))))).....)).((((.(((((((((...((((((((....)))).))))..))))))))).))))............... ( -37.70) >DroSim_CAF1 14586 120 + 1 UACGCUGGAGGGGGAGCAAAUGAAUCCGGAUUUCAUCAAGAUCAACCCGCAGCAUUCGAUUCCCACGCUGGUGGACAAUGGUUUCACCAUUUGGGAAUCGCGUGCCAUUCUCGUUUACCU ...((.(((((((((.........))).(((((.....)))))..)))...((((.(((((((((...(((((((.......)))))))..))))))))).))))...))).))...... ( -39.00) >DroEre_CAF1 14258 120 + 1 UACCCUGGAGGGGGAGCAACUGAAUCCGGAUUUCAUCAAGAUCAAUCCCCAGCACUCGAUUCCCACGCUGGUGGACAAUGGCUUCACCAUUUGGGAGUCGCGUGCCAUUCUGGUUUACCU ....(..((((((((.....(((.((..(((...)))..))))).))))).((((.(((((((((...(((((((.......)))))))..))))))))).))))..)))..)....... ( -44.80) >DroYak_CAF1 14303 120 + 1 UACCCUGGAAGGGGAGCAACUGAAUCCGGAUUUCAUCAAGAUCAACCCCCAGCACUCGAUUCCCACGCUGGUUGACAAUGGCUUUACCAUUUGGGAGUCUCGUGCCAUCCUGGUUUACCU .(((..(((.((((.(....(((.((..(((...)))..))))).))))).((((..((((((((...((((.............))))..))))))))..))))..))).)))...... ( -35.02) >consensus UACCCUGGAAGGGGAGCAAAUGAAUCCGGAUUUCAUCAAGAUCAACCCCCAGCACUCGAUUCCCACGCUGGUGGACAAUGGCUUCACCAUUUGGGAGUCGCGUGCCAUUCUCGUUUACCU .(((..(((.(((((.........))).(((((.....)))))...))...((((.(((((((((...(((((((.......)))))))..))))))))).))))..))).)))...... (-35.40 = -34.90 + -0.50)

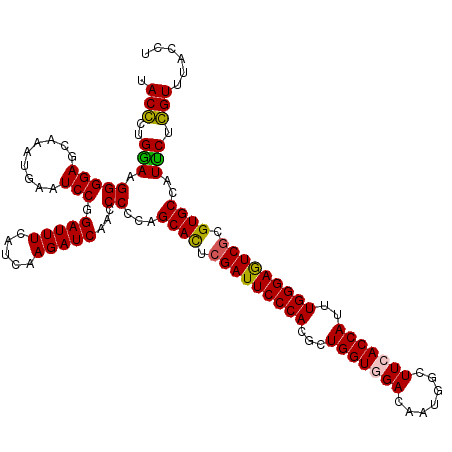
| Location | 8,198,898 – 8,199,018 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -41.10 |
| Consensus MFE | -33.46 |
| Energy contribution | -34.40 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8198898 120 - 27905053 AGGUAAACGAGAAUGGCACGCGACUCCCAAAUGGUGAAACCAUUGUCUACCAGCGUGGGAAUCGAGUGCUGCGGGUUGAUCUUGAUGAAAUCGGGAUUCAUUUGCUCCCCUUUCAGCGUA ........((((..(((((.(((.(((((..((((((........)).))))...))))).))).)))))((((((.(((((((((...))))))))).)))))).....))))...... ( -40.90) >DroSim_CAF1 14586 120 - 1 AGGUAAACGAGAAUGGCACGCGAUUCCCAAAUGGUGAAACCAUUGUCCACCAGCGUGGGAAUCGAAUGCUGCGGGUUGAUCUUGAUGAAAUCCGGAUUCAUUUGCUCCCCCUCCAGCGUA ........(((...((((..(((((((((..(((((...........)))))...)))))))))..))))((((((.(((((.(((...))).))))).)))))).....)))....... ( -40.70) >DroEre_CAF1 14258 120 - 1 AGGUAAACCAGAAUGGCACGCGACUCCCAAAUGGUGAAGCCAUUGUCCACCAGCGUGGGAAUCGAGUGCUGGGGAUUGAUCUUGAUGAAAUCCGGAUUCAGUUGCUCCCCCUCCAGGGUA ......(((.....(((((.(((.(((((..(((((...........)))))...))))).))).)))))((((((((((((.(((...))).))).))))....)))))......))). ( -42.30) >DroYak_CAF1 14303 120 - 1 AGGUAAACCAGGAUGGCACGAGACUCCCAAAUGGUAAAGCCAUUGUCAACCAGCGUGGGAAUCGAGUGCUGGGGGUUGAUCUUGAUGAAAUCCGGAUUCAGUUGCUCCCCUUCCAGGGUA ......(((.(((.(((((..((.(((((..((((...((....))..))))...))))).))..)))))((((((.(((((.(((...))).))))).....))))))..)))..))). ( -40.50) >consensus AGGUAAACCAGAAUGGCACGCGACUCCCAAAUGGUGAAACCAUUGUCCACCAGCGUGGGAAUCGAGUGCUGCGGGUUGAUCUUGAUGAAAUCCGGAUUCAGUUGCUCCCCCUCCAGCGUA ......(((.....(((((.(((.(((((..(((((...........)))))...))))).))).)))))((((((((((((.(((...))).))).))))....)))))......))). (-33.46 = -34.40 + 0.94)

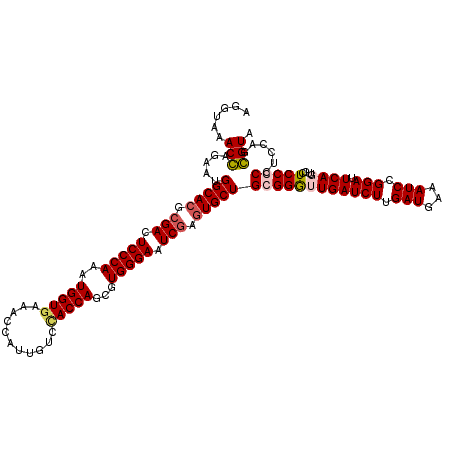
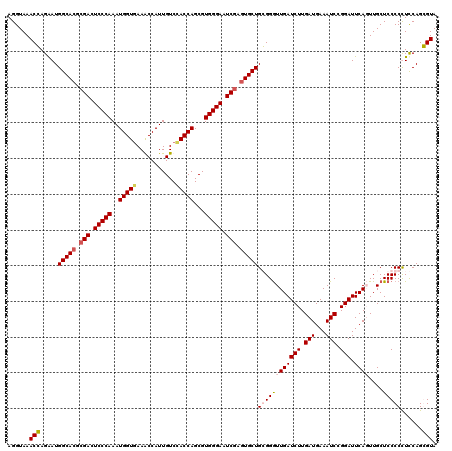
| Location | 8,198,938 – 8,199,058 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.06 |
| Mean single sequence MFE | -39.93 |
| Consensus MFE | -33.44 |
| Energy contribution | -33.62 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
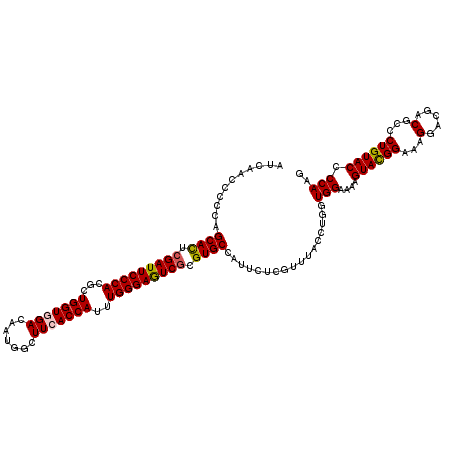
>3R_DroMel_CAF1 8198938 120 + 27905053 AUCAACCCGCAGCACUCGAUUCCCACGCUGGUAGACAAUGGUUUCACCAUUUGGGAGUCGCGUGCCAUUCUCGUUUACCUGGUGGAAAAGUACGGAAAGGAUGACGCGCUGUACCCCAAG ........(((((....((((((((...((((((((....)))).))))..))))))))((((.((.(((.(((...((....))......)))))).))...)))))))))........ ( -39.90) >DroSim_CAF1 14626 120 + 1 AUCAACCCGCAGCAUUCGAUUCCCACGCUGGUGGACAAUGGUUUCACCAUUUGGGAAUCGCGUGCCAUUCUCGUUUACCUGGUGGAAAAGUACGGAAAGGAUGACGCGCUGUACCCCAAG ........(((((....((((((((...(((((((.......)))))))..))))))))((((.((.(((.(((...((....))......)))))).))...)))))))))........ ( -41.70) >DroEre_CAF1 14298 120 + 1 AUCAAUCCCCAGCACUCGAUUCCCACGCUGGUGGACAAUGGCUUCACCAUUUGGGAGUCGCGUGCCAUUCUGGUUUACCUGGUGGAAAAGUACGGAAAGGACGACUCCCUGUACCCCAAG .....((((((((((.(((((((((...(((((((.......)))))))..))))))))).))))......((....))))).)))...((((((..((.....))..))))))...... ( -45.20) >DroYak_CAF1 14343 120 + 1 AUCAACCCCCAGCACUCGAUUCCCACGCUGGUUGACAAUGGCUUUACCAUUUGGGAGUCUCGUGCCAUCCUGGUUUACCUGGUGGAAAAGUAUGGAAAGGACGACUCCCUGUACCCCAAG .((((((...(((.............))))))))).(((((.....))))).(((((((((.(.((((.((..(((((...)))))..)).))))..).)).)))))))........... ( -32.92) >consensus AUCAACCCCCAGCACUCGAUUCCCACGCUGGUGGACAAUGGCUUCACCAUUUGGGAGUCGCGUGCCAUUCUCGUUUACCUGGUGGAAAAGUACGGAAAGGACGACGCCCUGUACCCCAAG ...........((((.(((((((((...(((((((.......)))))))..))))))))).)))).................(((....((((((...(.....)...)))))).))).. (-33.44 = -33.62 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:19 2006