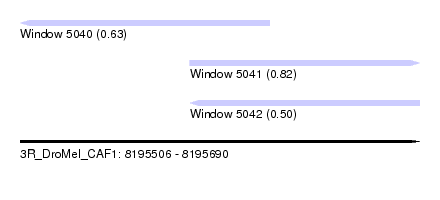
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,195,506 – 8,195,690 |
| Length | 184 |
| Max. P | 0.815535 |

| Location | 8,195,506 – 8,195,621 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.07 |
| Mean single sequence MFE | -27.55 |
| Consensus MFE | -18.14 |
| Energy contribution | -18.32 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8195506 115 - 27905053 AAAAGA---AUUUAUGUUUUUUCAGGCGCACAGAUGUUCAUUAGAAAUGCAAAAUUGUGUUUCA-AGCUAAUCGAUUGCA-UGCUUAUGUGUUUGCUAAUCUUAGCAAGUCGAUGUGUAA .(((((---((....)))))))...(((((..(((........((((((((....)))))))).-.(((((..(((((((-.((......)).))).)))))))))..)))..))))).. ( -27.50) >DroSim_CAF1 9776 115 - 1 AAAAGAAGAAUUUA----UUUUCAGGCGCGCAGAUGUUCAUUAGAAAUGCAAAAAUGUGUUUCA-AGCAAAUCGAGUGCACUGCGCAUGUGUUUGCCAACUUUAGCAAGUCGAAGUGUAA ....(((((.....----))))).(((((((((..........((((((((....)))))))).-.(((.......))).))))))......((((........)))))))......... ( -28.00) >DroEre_CAF1 11029 114 - 1 AGAGAGAGAAUUUA----UUUUCAGGCGCACAGAUGUUCAUAAGAAAUGCAAAAAUGUGUUUCAAAGCAAAUUGAGUGCAC--CAAAUGGGUGUGUCAGCUUUAGCAAGUCGAAGUGUAA .....(((((....----)))))..((((...(((........((((((((....))))))))(((((....(((.(((((--(.....)))))))))))))).....)))...)))).. ( -25.50) >DroYak_CAF1 9582 112 - 1 AUAAGAAGAAUUUA----UUUUCAGGCGCACAGAUGUUCAUUAGAAAUGCAAAAAUGUGUUUCA-AGCAAAUUGAGUGCAC--CAAAUGGGUGUGCCAACU-UAGCAAGUCGAGGUGUUA ....(((((.....----))))).(((((...(((........((((((((....)))))))).-.((((.(((.(..(((--(.....))))..)))).)-).))..)))...))))). ( -29.20) >consensus AAAAGAAGAAUUUA____UUUUCAGGCGCACAGAUGUUCAUUAGAAAUGCAAAAAUGUGUUUCA_AGCAAAUCGAGUGCAC__CAAAUGGGUGUGCCAACUUUAGCAAGUCGAAGUGUAA ........................(((((((............((((((((....))))))))...(((.......)))...........))))))).(((((........))))).... (-18.14 = -18.32 + 0.19)



| Location | 8,195,584 – 8,195,690 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.36 |
| Mean single sequence MFE | -16.69 |
| Consensus MFE | -10.89 |
| Energy contribution | -11.33 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8195584 106 + 27905053 UGAACAUCUGUGCGCCUGAAAAAACAUAAAU---UCUUUUAAAAUGCAAAAAAAAAUAAGUA---------CAAACAAAUAAGCAGGCAAACAGAAACCAGAUAAGAAU--UCAAGCCGG ......(((((..(((((........((((.---...))))..................((.---------...)).......)))))..)))))..............--......... ( -13.90) >DroSim_CAF1 9855 104 + 1 UGAACAUCUGCGCGCCUGAAAA----UAAAUUCUUCUUUUAAAAUGCAAA---AAAUAAGUA---------CAAACAUACAAACAGGCAAACAGAAAGCAGAUAAGAAUUUUCAGGCCGG ..........((.(((((((((----(....(((.(((((....(((...---......(((---------......)))......)))....))))).))).....)))))))))))). ( -22.16) >DroEre_CAF1 11107 111 + 1 UGAACAUCUGUGCGCCUGAAAA----UAAAUUCUCUCUCUAAAAUGCAAA---AAGUAAGCAAAUAAAUAAUAAGUAAAUAAACAGGCAAACAGAAAGCAGAUAACAAU--UCAAGCCGG ......(((((..(((((....----..................(((...---......))).....((.....)).......)))))..)))))..............--......... ( -14.00) >consensus UGAACAUCUGUGCGCCUGAAAA____UAAAUUCUUCUUUUAAAAUGCAAA___AAAUAAGUA_________CAAACAAAUAAACAGGCAAACAGAAAGCAGAUAAGAAU__UCAAGCCGG ......(((((..(((((........((((.......))))...(((............))).....................)))))..)))))......................... (-10.89 = -11.33 + 0.45)

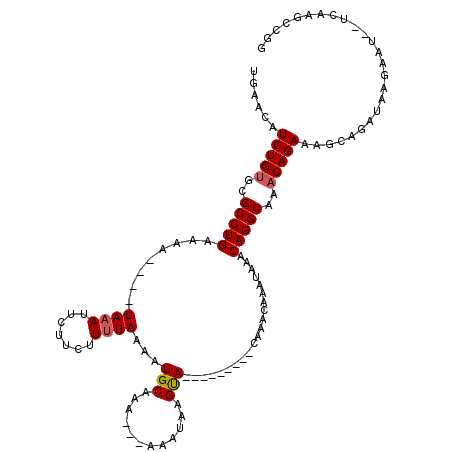
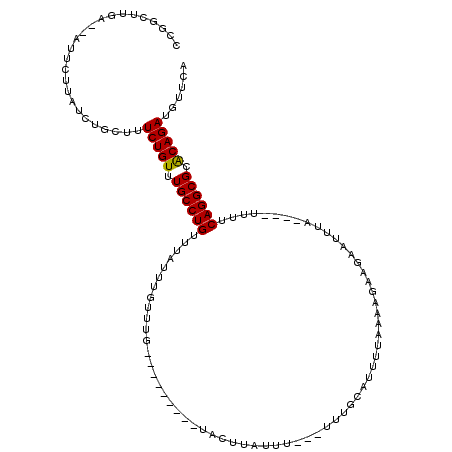
| Location | 8,195,584 – 8,195,690 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.36 |
| Mean single sequence MFE | -21.09 |
| Consensus MFE | -12.67 |
| Energy contribution | -12.45 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8195584 106 - 27905053 CCGGCUUGA--AUUCUUAUCUGGUUUCUGUUUGCCUGCUUAUUUGUUUG---------UACUUAUUUUUUUUUGCAUUUUAAAAGA---AUUUAUGUUUUUUCAGGCGCACAGAUGUUCA ((((..(((--....))).))))..(((((.((((((............---------.((.((..((((((((.....)))))))---)..)).)).....)))))).)))))...... ( -18.97) >DroSim_CAF1 9855 104 - 1 CCGGCCUGAAAAUUCUUAUCUGCUUUCUGUUUGCCUGUUUGUAUGUUUG---------UACUUAUUU---UUUGCAUUUUAAAAGAAGAAUUUA----UUUUCAGGCGCGCAGAUGUUCA .((((((((((((.....(((.((((..(..(((..((..((((....)---------)))..))..---...)))..)..)))).)))....)----))))))))).)).......... ( -24.20) >DroEre_CAF1 11107 111 - 1 CCGGCUUGA--AUUGUUAUCUGCUUUCUGUUUGCCUGUUUAUUUACUUAUUAUUUAUUUGCUUACUU---UUUGCAUUUUAGAGAGAGAAUUUA----UUUUCAGGCGCACAGAUGUUCA ..(((..((--.......)).))).(((((.((((((...............((((..(((......---...)))...))))...........----....)))))).)))))...... ( -20.11) >consensus CCGGCUUGA__AUUCUUAUCUGCUUUCUGUUUGCCUGUUUAUUUGUUUG_________UACUUAUUU___UUUGCAUUUUAAAAGAAGAAUUUA____UUUUCAGGCGCACAGAUGUUCA .........................(((((.((((((.................................................................)))))).)))))...... (-12.67 = -12.45 + -0.22)

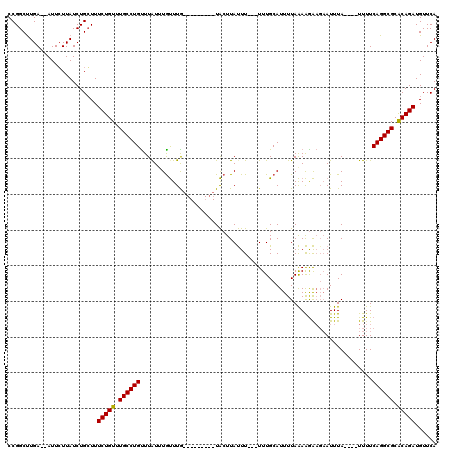
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:15 2006