
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,189,883 – 8,190,163 |
| Length | 280 |
| Max. P | 0.965134 |

| Location | 8,189,883 – 8,190,003 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -50.74 |
| Consensus MFE | -45.34 |
| Energy contribution | -45.46 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8189883 120 - 27905053 UGGCAGCCCGUAAUGGCAUUCGGGCCAUGGCCAACAGUGCUCUGGUGGGCUGUCUGGUGCUGGCCAUGAUUGAAGGAGCCGGUGCAGCAGUGGCCACCAUUAAUGCAGCCGACAAAGGUG .(((((((((..(.((((((..(((....)))...)))))).)..)))))))))(((((.(((((((..(((..(....)....)))..))))))).))))).....(((......))). ( -50.40) >DroSec_CAF1 4055 120 - 1 UGGCAGCCCGUAAUGGCAUUCGGGCCAUGGCCAACAGUGCUCUGGUGGGCUGUCUGGUGCUGGCCAUGAUUGAAGGAGCCGGUGCAGCAGUGGCCACCAUUAAUGCAGCCGACAAGGGUG ((((.(((((..........)))))....)))).....(((((.((.((((((.(((((.(((((((..(((..(....)....)))..))))))).)))))..)))))).)).))))). ( -51.90) >DroSim_CAF1 4069 120 - 1 UGGCAGCCCGCAAUGGCAUUCGGGCCAUGGCCAACAGUGCACUGGUGGGCUGUCUGGUGCUGGCCAUGAUUGAAGGAGCCGGUGCAGCAGUGGCCACCAUUAAUGCAGCCGACAAGGGUG .((((((((((....(((((..(((....)))...)))))....))))))))))(((((.(((((((..(((..(....)....)))..))))))).)))))...((.((.....)).)) ( -53.60) >DroEre_CAF1 3977 120 - 1 UGGCAGCCCGUAAUGGCAUUCGGGCCAUGGCCAACAGUGCUCUGGUGGGCUGCCUGGUUCUGGCCAUGAUUGAAGGAGCAGGUGCAGCAGUGGCCACCAUUAAUGCAGCCGAUAAGGGGG .(((((((((..(.((((((..(((....)))...)))))).)..)))))))))..(((((...((....))..))))).((((((..(((((...)))))..)))).)).......... ( -48.80) >DroYak_CAF1 4015 120 - 1 UGGCCGCUCGUAAUGGCUUUCGGGCCAUGGCCAACAGUGCUCUUGUGGGCUGCCUGGUCCUGGCCAUGAUUGAAGGAGCAGGUGCAGCAGUGGCCACCAUCAAUGCAGCUGAUAAAGGCC .((((((((.(.((((((...((((((.(((.......((((....)))).))))))))).))))))......).))))((.((((...((((...))))...)))).))......)))) ( -49.00) >consensus UGGCAGCCCGUAAUGGCAUUCGGGCCAUGGCCAACAGUGCUCUGGUGGGCUGUCUGGUGCUGGCCAUGAUUGAAGGAGCCGGUGCAGCAGUGGCCACCAUUAAUGCAGCCGACAAGGGUG ((((.(((((..........)))))....)))).....(((((.((.((((((.(((((.(((((((..(((............)))..))))))).)))))..)))))).)).))))). (-45.34 = -45.46 + 0.12)



| Location | 8,189,923 – 8,190,043 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.75 |
| Mean single sequence MFE | -39.10 |
| Consensus MFE | -33.50 |
| Energy contribution | -33.90 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8189923 120 + 27905053 GGCUCCUUCAAUCAUGGCCAGCACCAGACAGCCCACCAGAGCACUGUUGGCCAUGGCCCGAAUGCCAUUACGGGCUGCCAAUAUCCCACCAGUGGCCGCUCCACUGAGUAUCGAGUUCCA (((.........(((((((((((.......((.(....).))..)))))))))))(((((..........))))).)))........((((((((.....)))))).))........... ( -39.50) >DroSec_CAF1 4095 120 + 1 GGCUCCUUCAAUCAUGGCCAGCACCAGACAGCCCACCAGAGCACUGUUGGCCAUGGCCCGAAUGCCAUUACGGGCUGCCAAUAUCCCACCCGUGGCCGCUCCACUGAGUAUCGAGUUCCA (((.........(((((((((((.......((.(....).))..)))))))))))(((((..........))))).)))......(((....)))..((((....))))........... ( -35.30) >DroSim_CAF1 4109 120 + 1 GGCUCCUUCAAUCAUGGCCAGCACCAGACAGCCCACCAGUGCACUGUUGGCCAUGGCCCGAAUGCCAUUGCGGGCUGCCAAUAUCCCACCAGUGGCCGCUCCACUGAGUAUCGAGUUCCA (((((.........((((.(((.((.(((((..((....))..))))).((.(((((......))))).))))))))))).......((((((((.....)))))).))...)))))... ( -40.40) >DroEre_CAF1 4017 120 + 1 UGCUCCUUCAAUCAUGGCCAGAACCAGGCAGCCCACCAGAGCACUGUUGGCCAUGGCCCGAAUGCCAUUACGGGCUGCCAAUAUCCCACCAGUGGCCGCUCCACUGAGUAUCGAGUUCCA .((((.........(((......)))((((((((.((((.......))))..(((((......)))))...))))))))........((((((((.....)))))).))...)))).... ( -40.20) >DroYak_CAF1 4055 120 + 1 UGCUCCUUCAAUCAUGGCCAGGACCAGGCAGCCCACAAGAGCACUGUUGGCCAUGGCCCGAAAGCCAUUACGAGCGGCCAAUAUCCCACCAGUGGCCGCUCCACUGAGAAUCGAGUUCCA .((((.(((....((((((((...(((((...........)).)))))))))))(((......))).....(((((((((............)))))))))......)))..)))).... ( -40.10) >consensus GGCUCCUUCAAUCAUGGCCAGCACCAGACAGCCCACCAGAGCACUGUUGGCCAUGGCCCGAAUGCCAUUACGGGCUGCCAAUAUCCCACCAGUGGCCGCUCCACUGAGUAUCGAGUUCCA .((((.......(((((((((...(((...((.(....).)).))))))))))))(((((..........)))))..............((((((.....))))))......)))).... (-33.50 = -33.90 + 0.40)

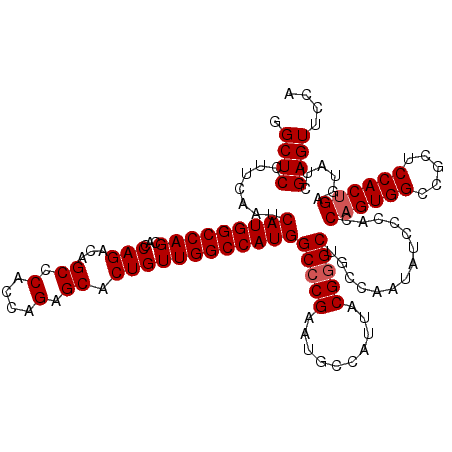
| Location | 8,189,923 – 8,190,043 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.75 |
| Mean single sequence MFE | -49.50 |
| Consensus MFE | -43.26 |
| Energy contribution | -43.50 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8189923 120 - 27905053 UGGAACUCGAUACUCAGUGGAGCGGCCACUGGUGGGAUAUUGGCAGCCCGUAAUGGCAUUCGGGCCAUGGCCAACAGUGCUCUGGUGGGCUGUCUGGUGCUGGCCAUGAUUGAAGGAGCC .((..((((((....(((((.....)))))(((.((.(((..((((((((..(.((((((..(((....)))...)))))).)..))))))).)..))))).)))...)))).))...)) ( -48.30) >DroSec_CAF1 4095 120 - 1 UGGAACUCGAUACUCAGUGGAGCGGCCACGGGUGGGAUAUUGGCAGCCCGUAAUGGCAUUCGGGCCAUGGCCAACAGUGCUCUGGUGGGCUGUCUGGUGCUGGCCAUGAUUGAAGGAGCC .((..((((((.....((((.....)))).(((.((.(((..((((((((..(.((((((..(((....)))...)))))).)..))))))).)..))))).)))...)))).))...)) ( -47.60) >DroSim_CAF1 4109 120 - 1 UGGAACUCGAUACUCAGUGGAGCGGCCACUGGUGGGAUAUUGGCAGCCCGCAAUGGCAUUCGGGCCAUGGCCAACAGUGCACUGGUGGGCUGUCUGGUGCUGGCCAUGAUUGAAGGAGCC .((..((((((....(((((.....)))))(((.((.(((..(((((((((....(((((..(((....)))...)))))....)))))))).)..))))).)))...)))).))...)) ( -51.40) >DroEre_CAF1 4017 120 - 1 UGGAACUCGAUACUCAGUGGAGCGGCCACUGGUGGGAUAUUGGCAGCCCGUAAUGGCAUUCGGGCCAUGGCCAACAGUGCUCUGGUGGGCUGCCUGGUUCUGGCCAUGAUUGAAGGAGCA ......(((((....(((((.....)))))(((.(((....(((((((((..(.((((((..(((....)))...)))))).)..)))))))))....))).)))...)))))....... ( -50.50) >DroYak_CAF1 4055 120 - 1 UGGAACUCGAUUCUCAGUGGAGCGGCCACUGGUGGGAUAUUGGCCGCUCGUAAUGGCUUUCGGGCCAUGGCCAACAGUGCUCUUGUGGGCUGCCUGGUCCUGGCCAUGAUUGAAGGAGCA ........(.((((((((.(((((((((.((......)).)))))))))...((((((...((((((.(((.......((((....)))).))))))))).)))))).)))).)))).). ( -49.70) >consensus UGGAACUCGAUACUCAGUGGAGCGGCCACUGGUGGGAUAUUGGCAGCCCGUAAUGGCAUUCGGGCCAUGGCCAACAGUGCUCUGGUGGGCUGUCUGGUGCUGGCCAUGAUUGAAGGAGCC .....(((.......(((((.....)))))(((.((.....((((((((((...((((((..(((....)))...))))))...)))))))))).....)).)))..........))).. (-43.26 = -43.50 + 0.24)



| Location | 8,190,043 – 8,190,163 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -40.74 |
| Consensus MFE | -34.12 |
| Energy contribution | -34.52 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8190043 120 + 27905053 GGCAUCCUCCCUCUGGCGAUAGUGGACCAGGGCGCAAUCCACUGUGCUGAAAGUGGCACCCCAAACGGCAAAGCUGCCGGCAAUGGACGGCGUUCUCAUCUUCACCAAAUCAAUGCCACC (((((........(((.((..(((....(((((((..((((..(((((......)))))......(((((....)))))....))))..))))))))))..)).))).....)))))... ( -41.22) >DroSec_CAF1 4215 120 + 1 GGCAUCCUCCCGCUGGCGAUAGUGGACCAGGGCGCAGUCCACUGUGCUGAAUGUGGCACCCCAAACGGCAAAGCUGCCGGCAAUGCACGGCGUUCUCAUCUUCACCAAAUCAAUGCCACC (((((....((((((....))))))....((((((((....)))))).((((((.(((.......(((((....)))))....)))...)))))).........))......)))))... ( -40.70) >DroSim_CAF1 4229 120 + 1 GGCAUCCUCCCGCUGGCGAUAGUGGACCAGGGCGCAGUCCACUGUGCUGAAAGUGGCACCCCAAACGGCAAAGCUGCCGGCAAUGCACGGCGUUCUCAUCUUCACCAAAUCAAUGCCCCC (((((....((((((....))))))....((((((((....))))))((((..(((.((.((...(((((....)))))((...))..)).)).).))..))))))......)))))... ( -39.90) >DroEre_CAF1 4137 120 + 1 GGCGUCCUCCCUCUGCCGAUAGUGGACCAGGACACAAUCCACUGUGCUGAACGUGGCUCCCCAAACGGCAAAGCUGCCGGCAAUGGAGGGCGUCCUCAUCUUCACCGAAUCAAUUCCACC ((((((((((...(((((((((((((...(....)..)))))))).......(..(((..((....))...)))..))))))..)))))))))).......................... ( -43.40) >DroYak_CAF1 4175 120 + 1 UGCAUCCUCCCUCUGGCGAUAGUGGACCAGGACGCAAUCCACUGUGCUGAAAGUGGCUCCCCAAACGGCAAAGCUGCCGGCAAUGGAGGGCGUUCUCAUCUUCACCGAAUCAAUGCCACC .............(((((((.(((((..(((((((...(((((........)))))((((.....(((((....))))).....)))).)))))))....)))))...)))...)))).. ( -38.50) >consensus GGCAUCCUCCCUCUGGCGAUAGUGGACCAGGGCGCAAUCCACUGUGCUGAAAGUGGCACCCCAAACGGCAAAGCUGCCGGCAAUGGACGGCGUUCUCAUCUUCACCAAAUCAAUGCCACC (((((........(((.((..(((....(((((((..((((.((((((......)))........(((((....)))))))).))))..))))))))))..)).))).....)))))... (-34.12 = -34.52 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:05 2006