
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,172,638 – 8,172,784 |
| Length | 146 |
| Max. P | 0.614994 |

| Location | 8,172,638 – 8,172,753 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.31 |
| Mean single sequence MFE | -21.95 |
| Consensus MFE | -19.23 |
| Energy contribution | -19.03 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
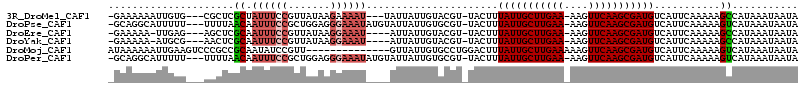
>3R_DroMel_CAF1 8172638 115 + 27905053 UGGUGUGCCCACUCACAUGUGAACAUUUAUUGGAAAACAUUAUUAUUUAUGGCUUUUUGAAUGACAUCGCUUGAACUU-UUCAAGCAAUAAAGUA-ACGUACAAUAAUA---AUUUUCUU .(((((...(((......))).)))))....(((((..((((((((((((((((((..((......))((((((....-.))))))...))))).-.)))).)))))))---))))))). ( -21.60) >DroVir_CAF1 150994 106 + 1 --GCGUGCGCGCUCAUAUGUGAACAUUUAUUGGAAAACAUUAUAAUUUAUGACUUUUUGAAUGACAUCGCUUGAACUUUUUCAAGCUAUAAAGUCCAGGGACAAUAAC------------ --(((....)))....(((....)))(((((.((((((((........)))..))))).)))))....(((((((....)))))))......(((....)))......------------ ( -20.10) >DroPse_CAF1 102910 116 + 1 --GUGUGCCCACUCAUAUGUGAACAUUUAUUGGAAAACAUUAUUAUUUAUGACUUUUUGAAUGACAUCGCUUGAACUU-UUCAAGCAAUAAAGUA-ACGCACAAUAAUACAUAUUUCCCU --.(((((.(((......))).((.((((((...........(((((((........)))))))....((((((....-.)))))))))))))).-..)))))................. ( -21.10) >DroGri_CAF1 134159 105 + 1 --GCGUGCGCGCUCAUAUGUGAACAUUUAUUGGAAAACAUUAUUAUUUAUGACUUUUUGAAUGACAUCGCUUGAACUU-UUCAAGCAAUAAAGUCCAGGCACAAUAAC------------ --..((((.(((......)))........(((((........(((((((........)))))))....((((((....-.)))))).......)))))))))......------------ ( -23.50) >DroMoj_CAF1 138070 106 + 1 --GCGUGCGCGCUCAUAUGUGAACAUUUAUUGGAAAACAUUAUUAUUUAUGACUUUUUGAAUGACAUCGCUUGAACUUUUUCAAGCAAUAAAGUCCAGGCACAAUAAC------------ --..((((.(((......)))........(((((........(((((((........)))))))....(((((((....))))))).......)))))))))......------------ ( -24.30) >DroPer_CAF1 104283 116 + 1 --GUGUGCCCACUCAUAUGUGAACAUUUAUUGGAAAACAUUAUUAUUUAUGACUUUUUGAAUGACAUCGCUUGAACUU-UUCAAGCAAUAAAGUA-ACGCACAAUAAUACAUAUUUCCCU --.(((((.(((......))).((.((((((...........(((((((........)))))))....((((((....-.)))))))))))))).-..)))))................. ( -21.10) >consensus __GCGUGCCCACUCAUAUGUGAACAUUUAUUGGAAAACAUUAUUAUUUAUGACUUUUUGAAUGACAUCGCUUGAACUU_UUCAAGCAAUAAAGUA_ACGCACAAUAAC____________ ....((((.(((......))).((..(((((.((((((((........)))..))))).)))))....(((((((....)))))))......))....)))).................. (-19.23 = -19.03 + -0.19)



| Location | 8,172,678 – 8,172,784 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.34 |
| Mean single sequence MFE | -22.08 |
| Consensus MFE | -13.14 |
| Energy contribution | -13.83 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.614994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8172678 106 - 27905053 -GAAAAAAUUGUG---CGCUCGCUAUUUCCGUUAUAAGAAAAU---UAUUAUUGUACGU-UACUUUAUUGCUUGAA-AAGUUCAAGCGAUGUCAUUCAAAAAGCCAUAAAUAAUA -.......(((((---.(((.........(((.((((......---..))))...))).-.....((((((((((.-....))))))))))..........))))))))...... ( -18.60) >DroPse_CAF1 102948 109 - 1 -GCAGGCAUUUUU---UUUUAACAAUUUCCGCUGGAGGGAAAUAUGUAUUAUUGUGCGU-UACUUUAUUGCUUGAA-AAGUUCAAGCGAUGUCAUUCAAAAAGUCAUAAAUAAUA -...(((......---...((((.((((((.......))))))..((((....))))))-))...((((((((((.-....))))))))))...........))).......... ( -22.50) >DroEre_CAF1 67101 104 - 1 -GAAAAA-UUGAG---AGCUCGCAAUUUCCGUUAUAAGGAAAU----AUUAUUGUACGU-UACUUUAUUGCUUGAA-AAGUUCAAGCGAUGUCAUUCAAAAAGCCAUAAAUAAUA -......-(((((---(((..(((((((((.......))))..----...)))))..))-)....((((((((((.-....))))))))))...)))))................ ( -19.40) >DroYak_CAF1 66125 104 - 1 -GAAAAA-AUGCG---AACUCGCAAUUUCCGUUAUAAGGAAAU----AUUAUUGUACGU-UACUUUAUUGCUUGAA-AAGUUCAAGCGAUGUCAUUCAAAAAGCCAUAAAUAAUA -(((...-.((((---....))))((((((.......))))))----............-.....((((((((((.-....))))))))))...))).................. ( -21.70) >DroMoj_CAF1 138108 101 - 1 AUAAAAAAUUGAAGUCCCGCCGCAAUAUCCGUU--------------GUUAUUGUGCCUGGACUUUAUUGCUUGAAAAAGUUCAAGCGAUGUCAUUCAAAAAGUCAUAAAUAAUA ........(((((((((.(.(((((((......--------------..))))))).).))))..(((((((((((....)))))))))))...)))))................ ( -27.80) >DroPer_CAF1 104321 109 - 1 -GCAGGCAUUUUU---UUUUAACAAUUUCCGCUGGAGGGAAAUAUGUAUUAUUGUGCGU-UACUUUAUUGCUUGAA-AAGUUCAAGCGAUGUCAUUCAAAAAGUCAUAAAUAAUA -...(((......---...((((.((((((.......))))))..((((....))))))-))...((((((((((.-....))))))))))...........))).......... ( -22.50) >consensus _GAAAAAAUUGUG___AGCUCGCAAUUUCCGUUAUAAGGAAAU____AUUAUUGUACGU_UACUUUAUUGCUUGAA_AAGUUCAAGCGAUGUCAUUCAAAAAGCCAUAAAUAAUA .....................((.((((((.......))))))......................(((((((((((....)))))))))))...........))........... (-13.14 = -13.83 + 0.70)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:58 2006