
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,160,487 – 8,160,603 |
| Length | 116 |
| Max. P | 0.755943 |

| Location | 8,160,487 – 8,160,603 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.47 |
| Mean single sequence MFE | -33.68 |
| Consensus MFE | -18.67 |
| Energy contribution | -20.20 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
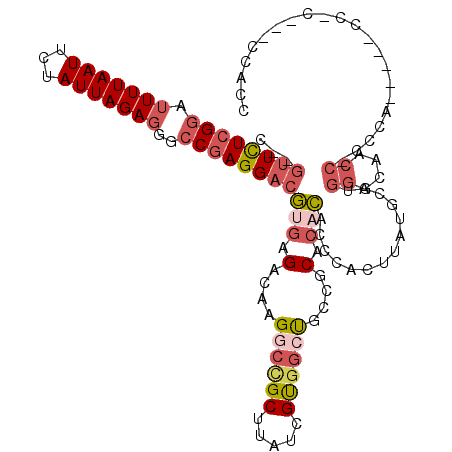
>3R_DroMel_CAF1 8160487 116 + 27905053 C--GUUCUCGGAUUUUAAUUCUAUUAGAGAGCCGAGGACGUGAGACAAGGCCGCUUAUCGUGGCUGCCGCACACACCCACUUAUGCAUGGGCAACCACCGAUUCGUCCAUACACCACC .--((((((((.(((((((...)))))))..))))))))(((.(....((((((.....))))))....).))).........((.((((((............)))))).))..... ( -34.60) >DroGri_CAF1 119920 95 + 1 CGCUUUUUGGGAUUUUAAUUCUAUUAGA----CGAGGCCAC-AGGCU-GGCCGCUUAUCGCUGCCGGCACACAUGUCGCCUGCUGC-----UAACA-UCCC---G-CC-C---CC--- ........(((((.(((..((.....))----..((((.((-(.(((-(((.((.....)).)))))).....))).)))).....-----))).)-))))---.-..-.---..--- ( -29.30) >DroSec_CAF1 54063 97 + 1 C--GUUCUCGGAUUUUAAUUCUAUUAGAGGGCCGAGGACGUGAGACAAGGCCGCUUAUCGUGGCUGCCGCACACACCCACUUAUGCAUGGGCAACC-------------------ACC .--((((((((.(((((((...)))))))..))))))))(((.(....((((((.....))))))....).))).............(((....))-------------------).. ( -34.20) >DroSim_CAF1 53633 97 + 1 C--GUUCUCGGAUUUUAAUUCUAUUAGAGGGCCGAGGACGUGAGACAAGUCUGCUUAUCGUGGCUGCCGCACACACCCACUUAUGCAUGGGCAACC-------------------ACC (--((((((((.(((((((...)))))))..)))))))))...((.(((....))).))((((.((((.((..((........))..)))))).))-------------------)). ( -31.80) >DroEre_CAF1 55129 113 + 1 C--GUUCUCGGAUUUUAAUUCUAUUAGAGGGCCGAGGACGUGAGACAAGGCCGCUUAUCGUGCCUGCCACACUCACCCACUUAUGCAUGGCUGGCCACCCAGUCACCCAC---CCACC .--((((((((.(((((((...)))))))..))))))))(((((...((((((.....)).))))......))))).......((..(((((((....)))))))..)).---..... ( -37.10) >DroYak_CAF1 54061 113 + 1 C--GUUCUCGGAUUUUAAUUCUAUUAGAGGGCCGAGGACGUGAGACAAGGCCGCUUAUCGUGGGUGCCGCACACACCCACUUAUGCAUGGCUAACCACCCAUUCACCCAC---CCGCC (--((((((((.(((((((...)))))))..)))))))))........(((.((.....(((((((.......)))))))....))((((........))))........---..))) ( -35.10) >consensus C__GUUCUCGGAUUUUAAUUCUAUUAGAGGGCCGAGGACGUGAGACAAGGCCGCUUAUCGUGGCUGCCGCACACACCCACUUAUGCAUGGGCAACC_CCCA_____CC_C___CCACC ...((((((((.(((((((...)))))))..))))))))(((.(....((((((.....))))))....).)))..............((....))...................... (-18.67 = -20.20 + 1.53)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:53 2006