
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,139,375 – 8,139,512 |
| Length | 137 |
| Max. P | 0.998699 |

| Location | 8,139,375 – 8,139,485 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 93.13 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -27.10 |
| Energy contribution | -27.30 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8139375 110 + 27905053 GAAUUGGAGGGCCGGAGGA---GGUUACUGGCCAACGAGCAGCCCAUAAACAGAGGACACAACAAAGUUGUUACAACAACAGCAUCGCCCGGUCACAAAAGUGACCAAAAUCA ((...((..((((((....---.....))))))..(((((.((((.......).)).)........(((((....))))).)).))).))((((((....))))))....)). ( -30.50) >DroSec_CAF1 32999 113 + 1 GAAUUGGAGGGCCGGAGGAGGAUGUUACUGGCCAACGAGCAGCCCAUAAACAGAGGACACAACAAAGUUGUUACAACAACAGCAUCGCCCGGUCACAAAAGUGACCAAAAUCA ((...((..((((((..(......)..))))))..(((((.((((.......).)).)........(((((....))))).)).))).))((((((....))))))....)). ( -30.40) >DroSim_CAF1 35382 113 + 1 GAAUUGGAGGGCAGGAGGAGGAGGUUACUGGCCAACGAGCAGCCCAUGAACAGAGGACACAACAAAGUUGUUACAACAACAGCAUCGCCCGGUCACAAAAGUGACCAAAAUCA ......((((((.((..(.(..(((.....)))..)...)..))......................((((((.....))))))...))))((((((....))))))....)). ( -30.00) >DroEre_CAF1 32822 109 + 1 -AAUUGGAGGGCCCGCGGC---GGUUGCUGGCCAAGGAGCAGCCCAUAAACAGAGGACGCAACAAAGUUGUUACAACAACAGCAUCGCCCGGUCACAAAAGUGACCAAAAUCA -.....((((((..((((.---(((((((........)))))))).....(....).)))......((((((.....))))))...))))((((((....))))))....)). ( -36.20) >DroYak_CAF1 32285 110 + 1 AAAUUGGAGGGCCGGACGA---GGUUGUUGGCCAAGGAGCAUCCCAUAAACAGAGGACACAACAAAGUUGUUACAACAACAGCAUCGCCCGGUCACAAAAGUGACCAAAAUCA ......((((((..((.(.---.(((((((((......)).((((.......).))).(((((...)))))..)))))))..).))))))((((((....))))))....)). ( -31.90) >consensus GAAUUGGAGGGCCGGAGGA___GGUUACUGGCCAACGAGCAGCCCAUAAACAGAGGACACAACAAAGUUGUUACAACAACAGCAUCGCCCGGUCACAAAAGUGACCAAAAUCA ......((((((..........(((.....)))...((((...((.........))..........(((((....))))).)).))))))((((((....))))))....)). (-27.10 = -27.30 + 0.20)

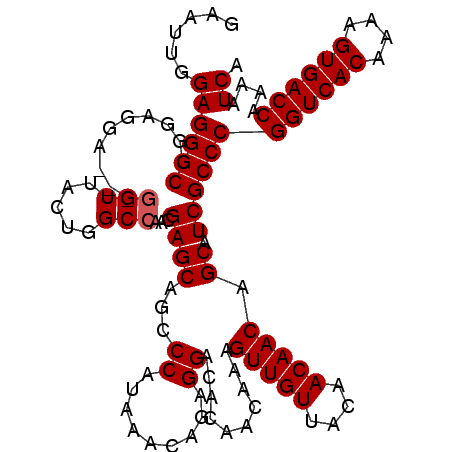

| Location | 8,139,375 – 8,139,485 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 93.13 |
| Mean single sequence MFE | -33.80 |
| Consensus MFE | -29.84 |
| Energy contribution | -29.88 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8139375 110 - 27905053 UGAUUUUGGUCACUUUUGUGACCGGGCGAUGCUGUUGUUGUAACAACUUUGUUGUGUCCUCUGUUUAUGGGCUGCUCGUUGGCCAGUAACC---UCCUCCGGCCCUCCAAUUC .......((((((....))))))(((((..((.(((((....)))))...))..))))).........((((((...(..((.......))---..)..))))))........ ( -32.10) >DroSec_CAF1 32999 113 - 1 UGAUUUUGGUCACUUUUGUGACCGGGCGAUGCUGUUGUUGUAACAACUUUGUUGUGUCCUCUGUUUAUGGGCUGCUCGUUGGCCAGUAACAUCCUCCUCCGGCCCUCCAAUUC .......((((((....))))))(((((((((.(((((....)))))......)))))...((((....((((.......))))...))))..........))))........ ( -32.20) >DroSim_CAF1 35382 113 - 1 UGAUUUUGGUCACUUUUGUGACCGGGCGAUGCUGUUGUUGUAACAACUUUGUUGUGUCCUCUGUUCAUGGGCUGCUCGUUGGCCAGUAACCUCCUCCUCCUGCCCUCCAAUUC .......((((((....))))))(((((((((.(((((....)))))......)))))....(((..(((.(........).)))..)))...........))))........ ( -30.40) >DroEre_CAF1 32822 109 - 1 UGAUUUUGGUCACUUUUGUGACCGGGCGAUGCUGUUGUUGUAACAACUUUGUUGCGUCCUCUGUUUAUGGGCUGCUCCUUGGCCAGCAACC---GCCGCGGGCCCUCCAAUU- .......((((((....))))))(((((..((.(((((....)))))...))..)))))........((((..((.((.((((........---)))).))))..))))...- ( -37.30) >DroYak_CAF1 32285 110 - 1 UGAUUUUGGUCACUUUUGUGACCGGGCGAUGCUGUUGUUGUAACAACUUUGUUGUGUCCUCUGUUUAUGGGAUGCUCCUUGGCCAACAACC---UCGUCCGGCCCUCCAAUUU .......((((((....))))))((((((((..(((((((.((((....))))(((((((........)))))))........))))))).---.))))..))))........ ( -37.00) >consensus UGAUUUUGGUCACUUUUGUGACCGGGCGAUGCUGUUGUUGUAACAACUUUGUUGUGUCCUCUGUUUAUGGGCUGCUCGUUGGCCAGUAACC___UCCUCCGGCCCUCCAAUUC .......((((((....))))))(((((((((.(((((....)))))......)))))....(((....((((.......))))...)))...........))))........ (-29.84 = -29.88 + 0.04)

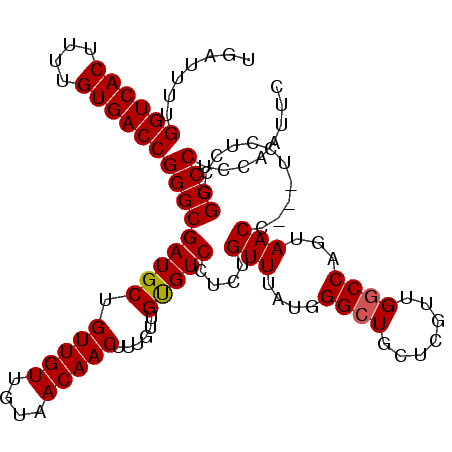

| Location | 8,139,411 – 8,139,512 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 94.32 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -22.57 |
| Energy contribution | -23.57 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.54 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.19 |
| SVM RNA-class probability | 0.998699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8139411 101 + 27905053 CAGCCCAUAAACAGAGGACACAACAAAGUUGUUACAACAACAGCAUCGCCCGGUCACAAAAGUGACCAAAAUCAGAGGCUACCAAAGUAGUAACAGAUGCU ...........................(((((....)))))((((((....((((((....)))))).......(..(((((....)))))..).)))))) ( -26.60) >DroSec_CAF1 33038 101 + 1 CAGCCCAUAAACAGAGGACACAACAAAGUUGUUACAACAACAGCAUCGCCCGGUCACAAAAGUGACCAAAAUCAGAGGCUACCAAAGUAGUAACAGAUGCU ...........................(((((....)))))((((((....((((((....)))))).......(..(((((....)))))..).)))))) ( -26.60) >DroSim_CAF1 35421 101 + 1 CAGCCCAUGAACAGAGGACACAACAAAGUUGUUACAACAACAGCAUCGCCCGGUCACAAAAGUGACCAAAAUCAGAGGCUACCAAAGUAGUAACAGAUGCU ...........................(((((....)))))((((((....((((((....)))))).......(..(((((....)))))..).)))))) ( -26.60) >DroEre_CAF1 32857 101 + 1 CAGCCCAUAAACAGAGGACGCAACAAAGUUGUUACAACAACAGCAUCGCCCGGUCACAAAAGUGACCAAAAUCAGAGGCUACCAAAGUAGUAACAGAUGCU ..((((.........))..))......(((((....)))))((((((....((((((....)))))).......(..(((((....)))))..).)))))) ( -27.50) >DroYak_CAF1 32321 101 + 1 CAUCCCAUAAACAGAGGACACAACAAAGUUGUUACAACAACAGCAUCGCCCGGUCACAAAAGUGACCAAAAUCAGGGGCUACCAAAGUAGUAACAGAUGCU ..((((.......).))).........(((((....)))))((((((....((((((....)))))).......(..(((((....)))))..).)))))) ( -29.70) >DroAna_CAF1 29492 97 + 1 CAUCCCAUAAACAGAGGACACAACAAAGUUGUAGGAACAACAGGAACCUCUG----CAAAAGUGACCAAAAUCAGGGGCUACCAAAGUAGUAACAGAUGCU ...(((.....((((((..(((((...))))).(......).....))))))----......(((......))))))(((((....))))).......... ( -20.10) >consensus CAGCCCAUAAACAGAGGACACAACAAAGUUGUUACAACAACAGCAUCGCCCGGUCACAAAAGUGACCAAAAUCAGAGGCUACCAAAGUAGUAACAGAUGCU ...........................(((((....)))))((((((....((((((....)))))).......(..(((((....)))))..).)))))) (-22.57 = -23.57 + 1.00)

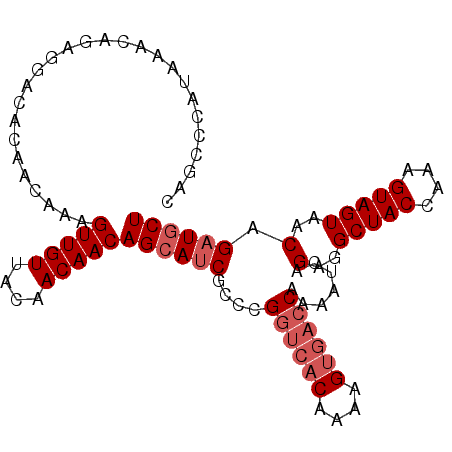

| Location | 8,139,411 – 8,139,512 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 94.32 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -25.61 |
| Energy contribution | -26.67 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8139411 101 - 27905053 AGCAUCUGUUACUACUUUGGUAGCCUCUGAUUUUGGUCACUUUUGUGACCGGGCGAUGCUGUUGUUGUAACAACUUUGUUGUGUCCUCUGUUUAUGGGCUG ((((((.((((((.....)))))).......(((((((((....))))))))).))))))(((((....)))))........((((.........)))).. ( -31.20) >DroSec_CAF1 33038 101 - 1 AGCAUCUGUUACUACUUUGGUAGCCUCUGAUUUUGGUCACUUUUGUGACCGGGCGAUGCUGUUGUUGUAACAACUUUGUUGUGUCCUCUGUUUAUGGGCUG ((((((.((((((.....)))))).......(((((((((....))))))))).))))))(((((....)))))........((((.........)))).. ( -31.20) >DroSim_CAF1 35421 101 - 1 AGCAUCUGUUACUACUUUGGUAGCCUCUGAUUUUGGUCACUUUUGUGACCGGGCGAUGCUGUUGUUGUAACAACUUUGUUGUGUCCUCUGUUCAUGGGCUG ....................((((((.(((....((((((....))))))(((((..((.(((((....)))))...))..))))).....))).)))))) ( -31.40) >DroEre_CAF1 32857 101 - 1 AGCAUCUGUUACUACUUUGGUAGCCUCUGAUUUUGGUCACUUUUGUGACCGGGCGAUGCUGUUGUUGUAACAACUUUGUUGCGUCCUCUGUUUAUGGGCUG ((((((.((((((.....)))))).......(((((((((....))))))))).))))))......((((((....))))))((((.........)))).. ( -33.30) >DroYak_CAF1 32321 101 - 1 AGCAUCUGUUACUACUUUGGUAGCCCCUGAUUUUGGUCACUUUUGUGACCGGGCGAUGCUGUUGUUGUAACAACUUUGUUGUGUCCUCUGUUUAUGGGAUG ((((((...((((.....))))((((........((((((....))))))))))))))))(((((....)))))........(((((........))))). ( -32.30) >DroAna_CAF1 29492 97 - 1 AGCAUCUGUUACUACUUUGGUAGCCCCUGAUUUUGGUCACUUUUG----CAGAGGUUCCUGUUGUUCCUACAACUUUGUUGUGUCCUCUGUUUAUGGGAUG .......((((((.....))))))(((((((....)))).....(----((((((.....(((((....)))))..........)))))))....)))... ( -22.86) >consensus AGCAUCUGUUACUACUUUGGUAGCCUCUGAUUUUGGUCACUUUUGUGACCGGGCGAUGCUGUUGUUGUAACAACUUUGUUGUGUCCUCUGUUUAUGGGCUG ((((((.((((((.....)))))).......(((((((((....))))))))).))))))(((((....)))))........((((.........)))).. (-25.61 = -26.67 + 1.06)


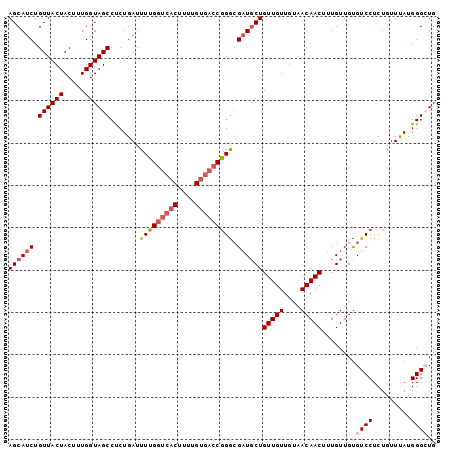
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:46 2006