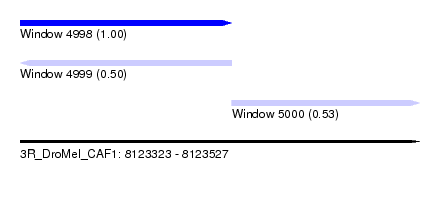
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,123,323 – 8,123,527 |
| Length | 204 |
| Max. P | 0.999805 |

| Location | 8,123,323 – 8,123,431 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 71.26 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -19.24 |
| Energy contribution | -19.48 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.65 |
| SVM decision value | 4.12 |
| SVM RNA-class probability | 0.999805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8123323 108 + 27905053 AAGCU-GCAAUUGCAUUUGAAUUGGCUGCAUGAGUCCAUAAACUGCAGCCGAUCAAUCGCUGCGCAGCCCAUACCCAAACCCAUACCCAUGCCCAUACCCAUGCCCAUA ..(((-((....((..((((.(((((((((.............)))))))))))))..))...)))))...................((((........))))...... ( -28.52) >DroVir_CAF1 63720 84 + 1 GCAGC-GCAAUUGCAUUUGAAUUGGCUGCAUGAGUCCAUAAACUGCAGCCGAUCAAUCGCUGCAAUGCCCAAGCCCC------------------------CUCCACCU (((((-((....))..((((.(((((((((.............)))))))))))))..)))))..............------------------------........ ( -27.32) >DroSec_CAF1 17012 84 + 1 AAGCU-GCAAUUGCAUUUGAAUUGGCUGCAUGAGUCCAUAAACUGCAGCCGAUCAAUCGCUGCGCAGCCCAUACCCA------------------------UACCCAUG ..(((-((....((..((((.(((((((((.............)))))))))))))..))...))))).........------------------------........ ( -27.42) >DroSim_CAF1 18266 84 + 1 AAGCU-GCAAUUGCAUUUGAAUUGGCUGCAUGAGUCCAUAAACUGCAGCCGAUCAAUCGCUGCGCAGCCCAUACCCA------------------------UACCCAUC ..(((-((....((..((((.(((((((((.............)))))))))))))..))...))))).........------------------------........ ( -27.42) >DroAna_CAF1 14451 107 + 1 UUGCUGGCAAUUGCAUUUGAAUUGGCUGCAUGAGUCCAUAAACUGCAGCCGAUCAAUCGCG-UUCUCCCCUGGCUCUGGCU-CUGGCUAUGGCUAUGGCUCUGGCUCCC ..((..(.....((..((((.(((((((((.............)))))))))))))..)).-.....((.(((((.((((.-...)))).))))).))..)..)).... ( -36.32) >consensus AAGCU_GCAAUUGCAUUUGAAUUGGCUGCAUGAGUCCAUAAACUGCAGCCGAUCAAUCGCUGCGCAGCCCAUACCCA________________________UACCCAUC ......(((...((..((((.(((((((((.............)))))))))))))..))))).............................................. (-19.24 = -19.48 + 0.24)



| Location | 8,123,323 – 8,123,431 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 71.26 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -19.32 |
| Energy contribution | -19.32 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8123323 108 - 27905053 UAUGGGCAUGGGUAUGGGCAUGGGUAUGGGUUUGGGUAUGGGCUGCGCAGCGAUUGAUCGGCUGCAGUUUAUGGACUCAUGCAGCCAAUUCAAAUGCAAUUGC-AGCUU ...((((.(((((...(((....(((((((((....((((((((((((..(((....))))).))))))))))))))))))).))).)))))..(((....))-))))) ( -35.70) >DroVir_CAF1 63720 84 - 1 AGGUGGAG------------------------GGGGCUUGGGCAUUGCAGCGAUUGAUCGGCUGCAGUUUAUGGACUCAUGCAGCCAAUUCAAAUGCAAUUGC-GCUGC .(.(.(((------------------------....))).).)...(((((..((((..((((((((((....)))...)))))))...))))..((....))-))))) ( -27.10) >DroSec_CAF1 17012 84 - 1 CAUGGGUA------------------------UGGGUAUGGGCUGCGCAGCGAUUGAUCGGCUGCAGUUUAUGGACUCAUGCAGCCAAUUCAAAUGCAAUUGC-AGCUU ((((....------------------------....))))(((((((..(((.((((..((((((((((....)))...)))))))...)))).)))...)))-)))). ( -29.10) >DroSim_CAF1 18266 84 - 1 GAUGGGUA------------------------UGGGUAUGGGCUGCGCAGCGAUUGAUCGGCUGCAGUUUAUGGACUCAUGCAGCCAAUUCAAAUGCAAUUGC-AGCUU ........------------------------.......((((((((..(((.((((..((((((((((....)))...)))))))...)))).)))...)))-))))) ( -29.10) >DroAna_CAF1 14451 107 - 1 GGGAGCCAGAGCCAUAGCCAUAGCCAG-AGCCAGAGCCAGGGGAGAA-CGCGAUUGAUCGGCUGCAGUUUAUGGACUCAUGCAGCCAAUUCAAAUGCAAUUGCCAGCAA .((..(..(.((..(.((....)).).-.))).)..)).((.((...-.(((.((((..((((((((((....)))...)))))))...)))).)))..)).))..... ( -26.40) >consensus GAUGGGCA________________________UGGGUAUGGGCUGCGCAGCGAUUGAUCGGCUGCAGUUUAUGGACUCAUGCAGCCAAUUCAAAUGCAAUUGC_AGCUU .................................................(((.((((..((((((((((....)))...)))))))...)))).)))............ (-19.32 = -19.32 + 0.00)


| Location | 8,123,431 – 8,123,527 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 85.90 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -22.91 |
| Energy contribution | -22.94 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.533917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8123431 96 + 27905053 CCCAUGCCCAUACCCAUGGUCUCCAGCUU-------GAAGAUUGUGGGAGACGAGACACGAUCGCGGUAGUAAAAUACCGGGUAUAGACUUACUAGAUCUUCU--- ...((((((...((((..((((.(.....-------).))))..)))).(.(((.......))))((((......))))))))))..................--- ( -28.50) >DroSec_CAF1 17096 96 + 1 CCCAUGCCCAUACCCAUGGUCUCCAGCUC-------GAAGAUUGUGGGAGACGAGACACGAUCGCGGUAGUAAAAUACCGGGUAUAGACUUACUAGACCUUCU--- ...((((((...((((..((((.(.....-------).))))..)))).(.(((.......))))((((......))))))))))..................--- ( -28.20) >DroSim_CAF1 18350 96 + 1 CCCAUGCCCAUACCCAUGGUCUCCAGCUC-------GAAGAUUGUGGGAGACGAGACACGAUCGCGGUAGUAAAAUACCGGGUAUAGACUUACUAGAUCUUCU--- ...((((((...((((..((((.(.....-------).))))..)))).(.(((.......))))((((......))))))))))..................--- ( -28.20) >DroEre_CAF1 16682 85 + 1 -----------ACCCAUGGUCUGCAGCUC-------GAAGAUUGUGGGAGACGAGACACGAUCGCGGUAGUAAAAUACCGGGUAUAGACUUACUAGAUCUUCU--- -----------.((((..((((.......-------..))))..))))...........((((.(((((......)))))((((......)))).))))....--- ( -26.70) >DroYak_CAF1 16205 85 + 1 -----------ACCCAGGGUCUUCAGCUC-------GAAGAUUGUGGGAGACGAGACACGAUCGCGGUAGUAAAAUACCGGGUAUAGACUUACUAGAUCUUCU--- -----------.((((.(((((((.....-------))))))).))))...........((((.(((((......)))))((((......)))).))))....--- ( -29.30) >DroPer_CAF1 45260 97 + 1 ---------GUCCCCAUCGUUUUCGACUCUUUCUUUGCAGAUUGUGGGAGACGAGACACGAUCGCGGUAGUAAAAUACCGGGUAUAGACUUACUAGAUCUUCUGCU ---------((((((((.((((.(((........))).)))).))))).))).(((...((((.(((((......)))))((((......)))).)))).)))... ( -26.90) >consensus _________AUACCCAUGGUCUCCAGCUC_______GAAGAUUGUGGGAGACGAGACACGAUCGCGGUAGUAAAAUACCGGGUAUAGACUUACUAGAUCUUCU___ ............((((..((((................))))..))))...........((((.(((((......)))))((((......)))).))))....... (-22.91 = -22.94 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:35 2006