
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,108,053 – 8,108,195 |
| Length | 142 |
| Max. P | 0.979100 |

| Location | 8,108,053 – 8,108,173 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.22 |
| Mean single sequence MFE | -29.74 |
| Consensus MFE | -12.72 |
| Energy contribution | -12.68 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8108053 120 + 27905053 ACCAGUACAUUAAUUGUGCUUAUGCCAUACAUGUCAAUGUAUCUAUUUGGUGCCAUCUUGAGGACCAUCACAUCUUGGCAAUUUUUAUUGCUGAGUAGCGAUCUCACCUCCAAGGUGCAA (((((((((.....))))).......((((((....)))))).....))))((.(((((((((...(((.(..((..(((((....)))))..))..).)))....))).)))))))).. ( -34.60) >DroSec_CAF1 1843 109 + 1 ACCAGUACGUUAGUUGUGUUUAUGCCAGACAUGUCAAUGUAUCUAUU-----------UGAGGACCACCGCAUCUUGGCAAUUUUUAUUGCUGAGUAGCGAUCUCACCUCCAAAGUGCAA ....(((((((.(.(((((((.....))))))).)))))))).....-----------.((((.....(((..((..(((((....)))))..))..)))......)))).......... ( -25.80) >DroSim_CAF1 1847 100 + 1 ACCAGUACAUUAGAUGUGUUUAU---------GUCAAUGUAUCUAUU-----------UGAGGACCAUCGCAUCUUGGCAAUUUUUAUUGCUGAGUAGCGAUCUCACCUCCAAGGUGCAA ....((((..((((((..((...---------...))..))))))..-----------.((((...(((((..((..(((((....)))))..))..)))))....))))....)))).. ( -31.90) >DroEre_CAF1 1390 112 + 1 AGCAGUACAUUGGUUA--CCUAAACCAUAAAUGCCAAAGUAUCGAA-UGGUGCCAUCCUGAGGAC-----UAUCUUGGCAAUUUUCAUUGCUGAGCAGCGAUCUCAGCGCCAAGGUGCAA .(((......(((((.--....)))))....)))....(((((...-((((((.....(((((.(-----...((..(((((....)))))..))....).))))))))))).))))).. ( -32.90) >DroYak_CAF1 810 91 + 1 AGCAAA-----------------------------AAUGUAACGAUUUGGUGCCAUCUUGAGGAGCAUCAUAUCUUGGCACUUUUUAUUGCUGAGUAGCGAUCUCACCGCCAAGGUGCAA .((...-----------------------------............(((((((........).)))))).((((((((.......((((((....))))))......)))))))))).. ( -23.52) >consensus ACCAGUACAUUAGUUGUGCUUAU_CCA_A_AUGUCAAUGUAUCUAUUUGGUGCCAUC_UGAGGACCAUCACAUCUUGGCAAUUUUUAUUGCUGAGUAGCGAUCUCACCUCCAAGGUGCAA ......................................................................((((((((........((((((....)))))).......))))))))... (-12.72 = -12.68 + -0.04)


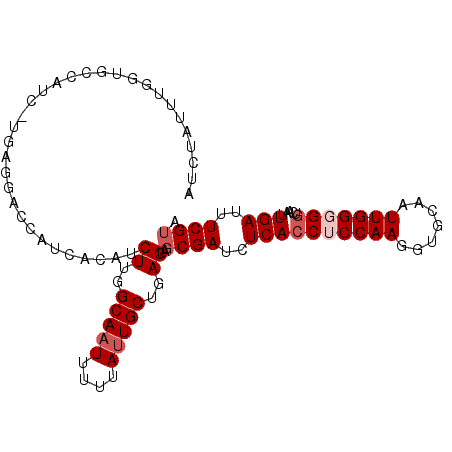
| Location | 8,108,093 – 8,108,195 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 84.84 |
| Mean single sequence MFE | -28.36 |
| Consensus MFE | -19.16 |
| Energy contribution | -20.16 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8108093 102 + 27905053 AUCUAUUUGGUGCCAUCUUGAGGACCAUCACAUCUUGGCAAUUUUUAUUGCUGAGUAGCGAUCUCACCUCCAAGGUGCAAUUGGGGGUCAAUUGAUUUCGUA .......((((.((.......))))))......((..(((((....)))))..))..((((..((((((((((.......)))))))).....))..)))). ( -29.30) >DroSec_CAF1 1883 91 + 1 AUCUAUU-----------UGAGGACCACCGCAUCUUGGCAAUUUUUAUUGCUGAGUAGCGAUCUCACCUCCAAAGUGCAAUUGGGGGUCAAUUGAUUUCGAA .....((-----------(((((.....(((..((..(((((....)))))..))..))).....((((((((.......)))))))).......))))))) ( -26.60) >DroSim_CAF1 1878 91 + 1 AUCUAUU-----------UGAGGACCAUCGCAUCUUGGCAAUUUUUAUUGCUGAGUAGCGAUCUCACCUCCAAGGUGCAAUUGGGGGUCAAUUGAUUUCGUA .......-----------.(((....(((((..((..(((((....)))))..))..))))))))((((((((.......)))))))).............. ( -28.70) >DroEre_CAF1 1428 96 + 1 AUCGAA-UGGUGCCAUCCUGAGGAC-----UAUCUUGGCAAUUUUCAUUGCUGAGCAGCGAUCUCAGCGCCAAGGUGCAAUUGGGGGUCAAUUGAUUUCGUA (((((.-((..(((..((..(....-----((((((((((((....)))((((((.......)))))))))))))))...)..))))))).)))))...... ( -28.20) >DroYak_CAF1 821 102 + 1 AACGAUUUGGUGCCAUCUUGAGGAGCAUCAUAUCUUGGCACUUUUUAUUGCUGAGUAGCGAUCUCACCGCCAAGGUGCAAUUGGGGGUCAAUUGAUUUCGUA ..((((((((((((........).))))))(((((((((.......((((((....))))))......))))))))).)))))(..(((....)))..)... ( -29.02) >consensus AUCUAUUUGGUGCCAUC_UGAGGACCAUCACAUCUUGGCAAUUUUUAUUGCUGAGUAGCGAUCUCACCUCCAAGGUGCAAUUGGGGGUCAAUUGAUUUCGUA .................................((..(((((....)))))..))..((((..((((((((((.......))))))).....)))..)))). (-19.16 = -20.16 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:29 2006