
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,104,268 – 8,104,524 |
| Length | 256 |
| Max. P | 0.999959 |

| Location | 8,104,268 – 8,104,376 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.48 |
| Mean single sequence MFE | -31.95 |
| Consensus MFE | -28.10 |
| Energy contribution | -28.34 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8104268 108 + 27905053 AGAGUAGUAUGGUGGGAGGGAGGGAGGCAA------------ACUGGGGCUUCUGUGGCAGCGAGUGCAAAAAGGCGCUCAAAUGGGGUUGUCAGUAUUUAACCCACAGGGGUUGGAGGU ...........(((((......((((((..------------......)))))).(((((((((((((......)))))).......)))))))........)))))............. ( -31.61) >DroSec_CAF1 56030 104 + 1 AGAGACGGAUGGCGGGAGG----GAGGGAA------------ACUGGGACUUCUGUGGCAGCGAGUGCAAAAAGGCGCUCAAAUGGGGUUGUCAGUAUUUAACCCACAGGGGUUGGAGGU ....((.....(((((((.----..((...------------.))....)))))))((((((((((((......)))))).......)))))).)).((((((((....))))))))... ( -35.31) >DroSim_CAF1 28094 104 + 1 AGAGUCGGUAGGUGGGAGG----GAGGGAA------------ACUGGGGCUUCUGUGGCAGCGAGUGCAAAAAGGCGCUCAAAUGGGGUUGUCAGUAUUUAACCCACAGGGGUUGGAGGU ...........(..((((.----..((...------------.))....))))..)((((((((((((......)))))).......))))))....((((((((....))))))))... ( -31.21) >DroEre_CAF1 30983 99 + 1 AGAGCCAGCAGGGGG--AG----GAGUGAA------------ACUGGGGCUUCUGUGGCAGCGAGUGCAAAAAGGCGCUCAAAUGGGGUUGUCAGUAUUUAACCCACAGGGGUUG---GU ...((((((..((((--((----((((...------------......)))))).(((((((((((((......)))))).......)))))))........))).)....))))---)) ( -32.11) >DroYak_CAF1 24475 114 + 1 AGAGUCAGCAGGGGG--AG----GAGGGAAAGGGAAAGGGGAACUGGGGCUUCUGUGGCAGCGAGUGCAAAAAGGCGCUCAAAUGGGGUUGUCAGUAUUUAACCCACAGGGGUUGGAGGU ...............--..----....................((..(((((((((......((((((......)))))).....((((((........)))))))))))))))..)).. ( -29.50) >consensus AGAGUCGGAAGGGGGGAGG____GAGGGAA____________ACUGGGGCUUCUGUGGCAGCGAGUGCAAAAAGGCGCUCAAAUGGGGUUGUCAGUAUUUAACCCACAGGGGUUGGAGGU ...........................................((..(((((((((......((((((......)))))).....((((((........)))))))))))))))..)).. (-28.10 = -28.34 + 0.24)

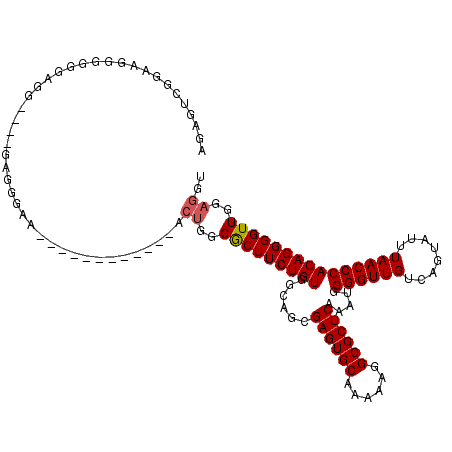

| Location | 8,104,298 – 8,104,416 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.64 |
| Mean single sequence MFE | -52.84 |
| Consensus MFE | -51.76 |
| Energy contribution | -52.00 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.61 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.88 |
| SVM RNA-class probability | 0.999959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8104298 118 + 27905053 --ACUGGGGCUUCUGUGGCAGCGAGUGCAAAAAGGCGCUCAAAUGGGGUUGUCAGUAUUUAACCCACAGGGGUUGGAGGUGCCGGCUGACUGCGCGAACUCCGCAGAAGUCCAUGUAACC --....((((((((((((.((.((((((......)))))).....(.((.((((((.((((((((....))))))))(....).)))))).)).)...))))))))))))))........ ( -52.80) >DroSec_CAF1 56056 118 + 1 --ACUGGGACUUCUGUGGCAGCGAGUGCAAAAAGGCGCUCAAAUGGGGUUGUCAGUAUUUAACCCACAGGGGUUGGAGGUGCCGGCUGACUGCGCGAACUCCGCAGAAGUCCAUGUAACC --....((((((((((((.((.((((((......)))))).....(.((.((((((.((((((((....))))))))(....).)))))).)).)...))))))))))))))........ ( -53.40) >DroSim_CAF1 28120 118 + 1 --ACUGGGGCUUCUGUGGCAGCGAGUGCAAAAAGGCGCUCAAAUGGGGUUGUCAGUAUUUAACCCACAGGGGUUGGAGGUGCCGGCUGACUGCGCGAACUCCGCAGAAGUCCAUGUAACC --....((((((((((((.((.((((((......)))))).....(.((.((((((.((((((((....))))))))(....).)))))).)).)...))))))))))))))........ ( -52.80) >DroEre_CAF1 31007 115 + 1 --ACUGGGGCUUCUGUGGCAGCGAGUGCAAAAAGGCGCUCAAAUGGGGUUGUCAGUAUUUAACCCACAGGGGUUG---GUGCCGGCUGACUGCGCGAACUCCGCAGAAGUCCAUGUAGCC --....((((((((((((.((.((((((......)))))).....(.((.((((((..(((((((....))))))---).....)))))).)).)...))))))))))))))........ ( -52.40) >DroYak_CAF1 24509 120 + 1 GAACUGGGGCUUCUGUGGCAGCGAGUGCAAAAAGGCGCUCAAAUGGGGUUGUCAGUAUUUAACCCACAGGGGUUGGAGGUGCCGGCUGACUGCGCGAACUCCGCAGAAGUCCAUGUAACC ......((((((((((((.((.((((((......)))))).....(.((.((((((.((((((((....))))))))(....).)))))).)).)...))))))))))))))........ ( -52.80) >consensus __ACUGGGGCUUCUGUGGCAGCGAGUGCAAAAAGGCGCUCAAAUGGGGUUGUCAGUAUUUAACCCACAGGGGUUGGAGGUGCCGGCUGACUGCGCGAACUCCGCAGAAGUCCAUGUAACC ......((((((((((((.((.((((((......)))))).....(.((.((((((.((((((((....)))))))).......)))))).)).)...))))))))))))))........ (-51.76 = -52.00 + 0.24)



| Location | 8,104,416 – 8,104,524 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.15 |
| Mean single sequence MFE | -20.55 |
| Consensus MFE | -16.67 |
| Energy contribution | -17.05 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8104416 108 - 27905053 CUAAUCCUGUGCUCCUCGCCACCUCUUGUUAGUGUGCCUCGCACUCGCAUCCAUGUUGGAGGUAACCCCAUCUCGAAACCCAGCC------------CCAUCCCUAAUCCUCAAUCCACU ......(((.((.....)).(((((((((.((((((...)))))).)))........)))))).................)))..------------....................... ( -15.60) >DroSec_CAF1 56174 95 - 1 CUAAUCCUGUGCUCCUCGCCACCUCUUGUUAGUGUUCCUCGCACUCGCAUCCAUGUUGGGGGUAAC-------------CCAGCC------------CCAACCCUAAUCCUCAAUCCACU ........(((...............(((.(((((.....))))).))).....(((((((.(...-------------..).))------------)))))..............))). ( -19.80) >DroEre_CAF1 31122 105 - 1 CUAAUCCUGUGCUCUGCGGCACCUCUUGUUAGAGUGCCUCGCACUCGCAUCCAUGUUGCGGGUAACCC---CUCAAAACCCAGCC------------CCAACCCUAAUCCUCAAUCCACU ........((((..((((((((.(((....))))))))..)))...))))....((((.((((.....---...........)))------------))))).................. ( -23.49) >DroYak_CAF1 24629 117 - 1 CUAAUCCUGUGCUCCGCGGCACCUCUUGUUAGUGUGCCUCGCACUCGCAUCCAUGUUGCGGGUAACCC---CUCGAAACCCAGCCCCAUCCCCAUCCCCAACCCUAAUGCUCAAUCCACU .......((.((...((((((.....(((.((((((...)))))).)))....))))))((((.....---......))))...........................)).))....... ( -23.30) >consensus CUAAUCCUGUGCUCCGCGCCACCUCUUGUUAGUGUGCCUCGCACUCGCAUCCAUGUUGCGGGUAACCC___CUCGAAACCCAGCC____________CCAACCCUAAUCCUCAAUCCACU .........(((.((((((((.....(((.(((((.....))))).)))....)))))))))))........................................................ (-16.67 = -17.05 + 0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:24 2006