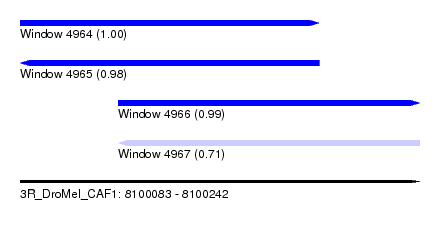
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,100,083 – 8,100,242 |
| Length | 159 |
| Max. P | 0.995959 |

| Location | 8,100,083 – 8,100,202 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.30 |
| Mean single sequence MFE | -46.66 |
| Consensus MFE | -39.56 |
| Energy contribution | -39.40 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.995959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8100083 119 + 27905053 CUUUGGGCAGCAUUGUGGUUUUGGCAA-UGUGGAAAGCGGUCGGCGAUUGAUGGCUAUCAAUGCCGAUGACUUAUAGCCAGAGUCGCGGACCGAGACUCACAUGAGUCGAGCCAGCCAAA .....(((.(((((((.......))))-)))((....(((((.((((((..(((((((((.......)))....)))))).)))))).))))).(((((....)))))...)).)))... ( -48.10) >DroSec_CAF1 51732 120 + 1 CUUUGGGAAGCUUUGUGGUCUUGGCAAGCGUGGAAAGCGGUCGGCGAUUGAUGGCUAUCAAUGCCGAUGACUUAUAGCCAGAGUCGCGGGCCGAGACUCACAUGAGUCGAGCCAGCCAAA ..((((...((((((((((((((((..((((((.(((..((((((.((((((....))))))))))))..)))....)))....)))..)))))))).))))......))))...)))). ( -48.30) >DroSim_CAF1 23803 119 + 1 CUUUGGGCAGCUUUGUGGUCUUGGCAA-CGUGGAAAGCGGUCGGCGAUUGAUGGCUAUCAAUGCCGAUGACUUAUAGCCGGAGUCGCGGACCGCGACUCACAUGAGUCGAGCCAGCCAAA .....(((.(((....)))..((((..-........((((((.((((((..(((((((((.......)))....)))))).)))))).))))))(((((....)))))..)))))))... ( -50.00) >DroEre_CAF1 26824 118 + 1 CUUUGGGCAACUUUGUGGUCUUGGCAA-CGUGGAAAGCGGUCGGCGAU-GAUGGCUAUCAAUGCCGAUGACUUAUAGCCAGAGUCGCAGACCGAGGCGCACAUGAGCAGAGCCAGCCAAA .....(((..((((((......(....-)((((....(((((.(((((-..(((((((((.......)))....))))))..))))).)))))...).)))....))))))...)))... ( -42.40) >DroYak_CAF1 20169 119 + 1 CUUUGGGCAACUUUGUGGUCUUGGCAA-CGUGGAAAGCGGUCGGCGAUUGAUGGCUAUCAAUGCCGAUGACUUAUAGCCGGAGUCGCAGACUCAGGCUCACAUGAGUCGAGCCAGCGAAA .....(((.....(((..(((((....-)).)))..)))((((((.((((((....))))))))))))(((((((((((.(((((...))))).))))...)))))))..)))....... ( -44.50) >consensus CUUUGGGCAGCUUUGUGGUCUUGGCAA_CGUGGAAAGCGGUCGGCGAUUGAUGGCUAUCAAUGCCGAUGACUUAUAGCCAGAGUCGCGGACCGAGACUCACAUGAGUCGAGCCAGCCAAA ...............((((..((((............(((((.(((((...(((((((((.......)))....))))))..))))).))))).(((((....)))))..)))))))).. (-39.56 = -39.40 + -0.16)



| Location | 8,100,083 – 8,100,202 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.30 |
| Mean single sequence MFE | -40.84 |
| Consensus MFE | -32.78 |
| Energy contribution | -33.62 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8100083 119 - 27905053 UUUGGCUGGCUCGACUCAUGUGAGUCUCGGUCCGCGACUCUGGCUAUAAGUCAUCGGCAUUGAUAGCCAUCAAUCGCCGACCGCUUUCCACA-UUGCCAAAACCACAAUGCUGCCCAAAG (((((..(((..(((((....))))).(((((.((((...(((((((..((.....))....)))))))....)))).))))).........-................)))..))))). ( -38.90) >DroSec_CAF1 51732 120 - 1 UUUGGCUGGCUCGACUCAUGUGAGUCUCGGCCCGCGACUCUGGCUAUAAGUCAUCGGCAUUGAUAGCCAUCAAUCGCCGACCGCUUUCCACGCUUGCCAAGACCACAAAGCUUCCCAAAG (((((..((((.......((((.((((.(((..(((....(((....((((..(((((((((((....)))))).)))))..)))).))))))..))).)))))))).))))..))))). ( -43.10) >DroSim_CAF1 23803 119 - 1 UUUGGCUGGCUCGACUCAUGUGAGUCGCGGUCCGCGACUCCGGCUAUAAGUCAUCGGCAUUGAUAGCCAUCAAUCGCCGACCGCUUUCCACG-UUGCCAAGACCACAAAGCUGCCCAAAG (((((..((((((((((....)))))).((((.(((((...((....((((..(((((((((((....)))))).)))))..)))).))..)-))))...))))....))))..))))). ( -45.60) >DroEre_CAF1 26824 118 - 1 UUUGGCUGGCUCUGCUCAUGUGCGCCUCGGUCUGCGACUCUGGCUAUAAGUCAUCGGCAUUGAUAGCCAUC-AUCGCCGACCGCUUUCCACG-UUGCCAAGACCACAAAGUUGCCCAAAG ...(((.(((...((......)))))..((((((((((..(((....((((..(((((..((((....)))-)..)))))..)))).))).)-))))..)))))........)))..... ( -38.70) >DroYak_CAF1 20169 119 - 1 UUUCGCUGGCUCGACUCAUGUGAGCCUGAGUCUGCGACUCCGGCUAUAAGUCAUCGGCAUUGAUAGCCAUCAAUCGCCGACCGCUUUCCACG-UUGCCAAGACCACAAAGUUGCCCAAAG ......(((..(((((..((((((((.(((((...))))).))))....(((.(((((((((((....)))))).)))))..((........-..))...))))))).))))).)))... ( -37.90) >consensus UUUGGCUGGCUCGACUCAUGUGAGUCUCGGUCCGCGACUCUGGCUAUAAGUCAUCGGCAUUGAUAGCCAUCAAUCGCCGACCGCUUUCCACG_UUGCCAAGACCACAAAGCUGCCCAAAG ((((((.(((((.((....))))))).(((((.((((...(((((((..((.....))....)))))))....)))).)))))............))))))................... (-32.78 = -33.62 + 0.84)



| Location | 8,100,122 – 8,100,242 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -41.06 |
| Consensus MFE | -37.18 |
| Energy contribution | -37.14 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8100122 120 + 27905053 UCGGCGAUUGAUGGCUAUCAAUGCCGAUGACUUAUAGCCAGAGUCGCGGACCGAGACUCACAUGAGUCGAGCCAGCCAAACUGGCAAAUCCAUCCAUCUAUGUGGCACAUUUGACUGCAG (((((.((((((....))))))))))).(((((.......)))))((((.....(((((....)))))..(((((((.....))).....(((......)))))))........)))).. ( -39.60) >DroSec_CAF1 51772 120 + 1 UCGGCGAUUGAUGGCUAUCAAUGCCGAUGACUUAUAGCCAGAGUCGCGGGCCGAGACUCACAUGAGUCGAGCCAGCCAAACUGGCAAAUCCAUCCAUCUAUGUGGCACAUUUGACUGCAG (((((.((((((....))))))))))).(((((.......)))))((((((((.(((((....)))))..(((((.....))))).................)))).(....).)))).. ( -41.50) >DroSim_CAF1 23842 120 + 1 UCGGCGAUUGAUGGCUAUCAAUGCCGAUGACUUAUAGCCGGAGUCGCGGACCGCGACUCACAUGAGUCGAGCCAGCCAAACUGGCAAAUCCAUCCAUCUAUGUGGCACAUUUGACUGCAG (((((.((((((....)))))))))))((((((((.(...((((((((...)))))))).))))))))).(((((.....)))))...............((..(((....)).)..)). ( -44.20) >DroEre_CAF1 26863 119 + 1 UCGGCGAU-GAUGGCUAUCAAUGCCGAUGACUUAUAGCCAGAGUCGCAGACCGAGGCGCACAUGAGCAGAGCCAGCCAAACUGGCAAAUCCAUCCAUCUAUGUGGCACAUUUGACUGCAG ((((((.(-(((....)))).)))))).(((((.......)))))((((.(.((.((.(((((((...(((((((.....))))).......))..)).)))))))...)).).)))).. ( -36.51) >DroYak_CAF1 20208 120 + 1 UCGGCGAUUGAUGGCUAUCAAUGCCGAUGACUUAUAGCCGGAGUCGCAGACUCAGGCUCACAUGAGUCGAGCCAGCGAAACUGGCAAAUCCAUCCAUCUAUGUGGCACAUUUGACUGCAG (((((.((((((....)))))))))))((((((((((((.(((((...))))).))))...)))))))).(((((.....)))))...............((..(((....)).)..)). ( -43.50) >consensus UCGGCGAUUGAUGGCUAUCAAUGCCGAUGACUUAUAGCCAGAGUCGCGGACCGAGACUCACAUGAGUCGAGCCAGCCAAACUGGCAAAUCCAUCCAUCUAUGUGGCACAUUUGACUGCAG (((((.((((((....))))))))))).(((((.......)))))((((.....(((((....)))))..(((((.....)))))........((((....)))).........)))).. (-37.18 = -37.14 + -0.04)

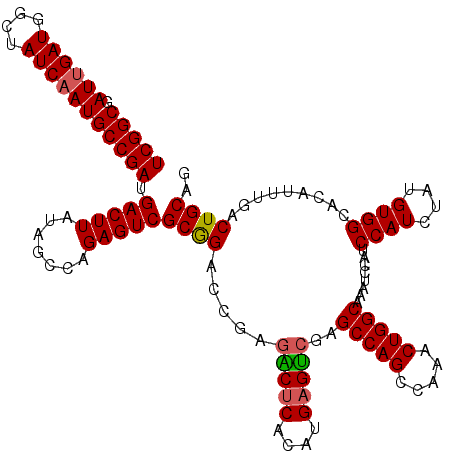

| Location | 8,100,122 – 8,100,242 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -41.40 |
| Consensus MFE | -34.84 |
| Energy contribution | -35.48 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8100122 120 - 27905053 CUGCAGUCAAAUGUGCCACAUAGAUGGAUGGAUUUGCCAGUUUGGCUGGCUCGACUCAUGUGAGUCUCGGUCCGCGACUCUGGCUAUAAGUCAUCGGCAUUGAUAGCCAUCAAUCGCCGA ....((((..(((.....))).(.(((((.((...(((((.....)))))..(((((....))))))).)))))))))).((((.....))))(((((((((((....)))))).))))) ( -41.90) >DroSec_CAF1 51772 120 - 1 CUGCAGUCAAAUGUGCCACAUAGAUGGAUGGAUUUGCCAGUUUGGCUGGCUCGACUCAUGUGAGUCUCGGCCCGCGACUCUGGCUAUAAGUCAUCGGCAUUGAUAGCCAUCAAUCGCCGA ...((..((..((((...))))..))..))((((((((((..(.((.((((.(((((....)))))..)))).)).)..)))))...))))).(((((((((((....)))))).))))) ( -43.80) >DroSim_CAF1 23842 120 - 1 CUGCAGUCAAAUGUGCCACAUAGAUGGAUGGAUUUGCCAGUUUGGCUGGCUCGACUCAUGUGAGUCGCGGUCCGCGACUCCGGCUAUAAGUCAUCGGCAUUGAUAGCCAUCAAUCGCCGA ...(((((((((...(((......))).(((.....))))))))))))....((((.((((((((((((...))))))))..)).)).)))).(((((((((((....)))))).))))) ( -43.10) >DroEre_CAF1 26863 119 - 1 CUGCAGUCAAAUGUGCCACAUAGAUGGAUGGAUUUGCCAGUUUGGCUGGCUCUGCUCAUGUGCGCCUCGGUCUGCGACUCUGGCUAUAAGUCAUCGGCAUUGAUAGCCAUC-AUCGCCGA ..((((.(....((((.(((....((..((((...(((((.....)))))))))..)))))))))....).))))((((.........)))).(((((..((((....)))-)..))))) ( -37.90) >DroYak_CAF1 20208 120 - 1 CUGCAGUCAAAUGUGCCACAUAGAUGGAUGGAUUUGCCAGUUUCGCUGGCUCGACUCAUGUGAGCCUGAGUCUGCGACUCCGGCUAUAAGUCAUCGGCAUUGAUAGCCAUCAAUCGCCGA .((.((((((((.(((((......))).)).))))(((((.....)))))..))))))....((((.(((((...))))).))))........(((((((((((....)))))).))))) ( -40.30) >consensus CUGCAGUCAAAUGUGCCACAUAGAUGGAUGGAUUUGCCAGUUUGGCUGGCUCGACUCAUGUGAGUCUCGGUCCGCGACUCUGGCUAUAAGUCAUCGGCAUUGAUAGCCAUCAAUCGCCGA ...((..((..((((...))))..))..))((((((((((..(.((.((((.(((((....)))))..)))).)).)..)))))...))))).(((((((((((....)))))).))))) (-34.84 = -35.48 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:04 2006