
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,099,732 – 8,099,979 |
| Length | 247 |
| Max. P | 0.982828 |

| Location | 8,099,732 – 8,099,849 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.70 |
| Mean single sequence MFE | -32.84 |
| Consensus MFE | -28.52 |
| Energy contribution | -28.92 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8099732 117 + 27905053 AUAUGGUUAACAACUCUUGACCAAGCAACGGUUAAAAAGGUUCAACGAGCCAUCGUCUUAGGCAGCGAAUGAGGCCCAAGACUUUUGGCUCUGCCAACUGACAAAGCGACUGACCAC--- ...((((((.......((((((.......))))))...(((.....((((((..(((((.(((..(....)..))).)))))...)))))).)))...............)))))).--- ( -34.80) >DroSec_CAF1 51377 117 + 1 AUAUGGUUAACAACUCUUGACCAAGCAACGGUUAAAAAGGUUCAACGAGCCAUCGUCUUAGCCAGCGAAUGAGGCCCCAGACUUUUGGCUCUGCCAACUGACAAAGCGACUGACCAC--- ...((((((.......((((((.......))))))...(((.....((((((..((((..(((.........)))...))))...)))))).)))...............)))))).--- ( -30.90) >DroSim_CAF1 23448 117 + 1 AUAUGGUUAACAACUCUUGACCAAGCAACGGUUAAAAAGGUUCAACGAGCCAUCGUCUUAGCCAGCGAAUGAGGCCCAAGACUUUUGGCUCUGCCAACUGACAAAGCGACUGACCAC--- ...((((((.......((((((.......))))))...(((.....((((((..(((((.(((.........)))..)))))...)))))).)))...............)))))).--- ( -32.30) >DroEre_CAF1 26442 120 + 1 AUAUGGUUAACAACUCUUGACCAAGCAACGGUUAAAAAGGUUCAACGAGCCAUCGUCUUCGCCAGCGAAUGAGGCCCAAGACUUUUGGCUCUGGCAACUGACAAAGUGACUGACCACCAC ...((((((...(((.((((((.......))))))...(((((...((((((..(((((.(((.........)))..)))))...))))))..).)))).....)))...)))))).... ( -32.40) >DroYak_CAF1 19804 117 + 1 AUAUGGUUAACAACUCUUGACCAAGCAACGGUUAAAAAGGUUCAACGAGCCAUCGUCUUUGCCAGCGAAUGAGGCCCAAGACUUUUGGCUCUGGCAACUGACAAAGUGACUGACCAC--- ...(((((((......)))))))......(((((....(((((...)))))...(((.(((((((.(...(((........)))....).)))))))..)))........)))))..--- ( -33.80) >consensus AUAUGGUUAACAACUCUUGACCAAGCAACGGUUAAAAAGGUUCAACGAGCCAUCGUCUUAGCCAGCGAAUGAGGCCCAAGACUUUUGGCUCUGCCAACUGACAAAGCGACUGACCAC___ ...((((((.......((((((.......))))))...((((....((((((..(((((.(((.........)))..)))))...))))))....))))...........)))))).... (-28.52 = -28.92 + 0.40)



| Location | 8,099,732 – 8,099,849 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.70 |
| Mean single sequence MFE | -37.79 |
| Consensus MFE | -32.54 |
| Energy contribution | -33.10 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8099732 117 - 27905053 ---GUGGUCAGUCGCUUUGUCAGUUGGCAGAGCCAAAAGUCUUGGGCCUCAUUCGCUGCCUAAGACGAUGGCUCGUUGAACCUUUUUAACCGUUGCUUGGUCAAGAGUUGUUAACCAUAU ---(((((.((..(((((.......((((((((((...(((((((((..(....)..)))))))))..))))))((((((....))))))...))))......)))))..)).))))).. ( -41.32) >DroSec_CAF1 51377 117 - 1 ---GUGGUCAGUCGCUUUGUCAGUUGGCAGAGCCAAAAGUCUGGGGCCUCAUUCGCUGGCUAAGACGAUGGCUCGUUGAACCUUUUUAACCGUUGCUUGGUCAAGAGUUGUUAACCAUAU ---(((((.((..(((((.......((((((((((...((((..((((.........)))).))))..))))))((((((....))))))...))))......)))))..)).))))).. ( -38.42) >DroSim_CAF1 23448 117 - 1 ---GUGGUCAGUCGCUUUGUCAGUUGGCAGAGCCAAAAGUCUUGGGCCUCAUUCGCUGGCUAAGACGAUGGCUCGUUGAACCUUUUUAACCGUUGCUUGGUCAAGAGUUGUUAACCAUAU ---(((((.((..(((((.......((((((((((...(((((((.((.........)))))))))..))))))((((((....))))))...))))......)))))..)).))))).. ( -38.42) >DroEre_CAF1 26442 120 - 1 GUGGUGGUCAGUCACUUUGUCAGUUGCCAGAGCCAAAAGUCUUGGGCCUCAUUCGCUGGCGAAGACGAUGGCUCGUUGAACCUUUUUAACCGUUGCUUGGUCAAGAGUUGUUAACCAUAU (((((...(((.(.(((..(((((.((..((((((...(((((.(.((.........))).)))))..))))))((((((....)))))).)).)).)))..))).))))...))))).. ( -35.10) >DroYak_CAF1 19804 117 - 1 ---GUGGUCAGUCACUUUGUCAGUUGCCAGAGCCAAAAGUCUUGGGCCUCAUUCGCUGGCAAAGACGAUGGCUCGUUGAACCUUUUUAACCGUUGCUUGGUCAAGAGUUGUUAACCAUAU ---(((((.((..(((((((((((.((..((((((...(((((..(((.........))).)))))..))))))((((((....)))))).)).)).))).).)))))..)).))))).. ( -35.70) >consensus ___GUGGUCAGUCGCUUUGUCAGUUGGCAGAGCCAAAAGUCUUGGGCCUCAUUCGCUGGCUAAGACGAUGGCUCGUUGAACCUUUUUAACCGUUGCUUGGUCAAGAGUUGUUAACCAUAU ...(((((.((..((((((((((.(..(.((((((...(((((((.((.........)))))))))..))))))((((((....)))))).)..).)))).).)))))..)).))))).. (-32.54 = -33.10 + 0.56)

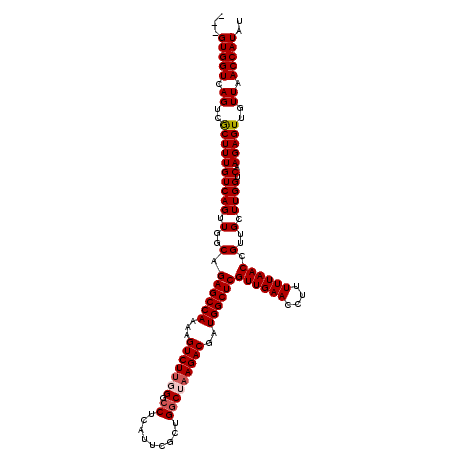
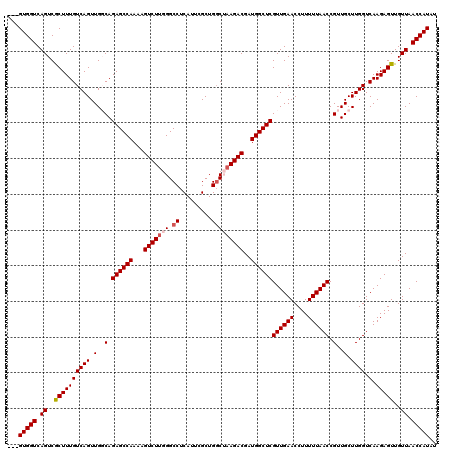
| Location | 8,099,812 – 8,099,907 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.19 |
| Mean single sequence MFE | -38.90 |
| Consensus MFE | -26.01 |
| Energy contribution | -26.17 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8099812 95 + 27905053 ACUUUUGGCUCUGCCAACUGACAAAGCGACUGACCAC---------------AAGUCAAAUUGGUGAACGGAGG-------GGCGCUGCGGUUGCCGAGGCAGCCGCC---GCCAAGAAA .((((((..((.(((((((.....)).((((......---------------.))))...))))))).))))))-------((((..((((((((....)))))))))---)))...... ( -36.80) >DroSec_CAF1 51457 95 + 1 ACUUUUGGCUCUGCCAACUGACAAAGCGACUGACCAC---------------AAGCCAAAUUGGUGGACAGAGG-------AGCGCUGCGGUUGCCGGGGCAGCCGCC---GCCAAGAAA ..(((((((...............((((.(((.((((---------------((......)).)))).)))...-------..))))((((((((....)))))))).---))))))).. ( -36.10) >DroSim_CAF1 23528 95 + 1 ACUUUUGGCUCUGCCAACUGACAAAGCGACUGACCAC---------------AAGCCAAAUUGAUGGACAGAGG-------GGAGCUGCGGUUGCCGAGGCAGCCGCC---GCCAAGAAA ..(((((((.((.((..(((.((.......)).((((---------------((......))).))).)))..)-------).))..((((((((....)))))))).---))))))).. ( -35.20) >DroEre_CAF1 26522 120 + 1 ACUUUUGGCUCUGGCAACUGACAAAGUGACUGACCACCACCACAAGCCAGCCAAGCCGAAUUGGUGGACCGAGGACUGGUGGACUCUGCGGUUACCGAGGCAGCCGCCGUCGCCAAGAAA ..(((((((..((((..(((.....(((((((.((((((((.(...(((.((((......)))))))...).))..))))))......))))))).....)))..))))..))))))).. ( -43.70) >DroYak_CAF1 19884 95 + 1 ACUUUUGGCUCUGGCAACUGACAAAGUGACUGACCAC---------------AAGCCAAAUCGGUGGACCGAGG-------GGCUCUGCGGUUGCCGAGGCAGCCGCC---GCCAAGAAA ..(((((((..(((((((((.((..(((......)))---------------.((((...((((....))))..-------)))).))))))))))).(((....)))---))))))).. ( -42.70) >consensus ACUUUUGGCUCUGCCAACUGACAAAGCGACUGACCAC_______________AAGCCAAAUUGGUGGACAGAGG_______GGCGCUGCGGUUGCCGAGGCAGCCGCC___GCCAAGAAA ..(((((((((((...(((((....((...........................))....)))))...))))...............((((((((....))))))))....))))))).. (-26.01 = -26.17 + 0.16)



| Location | 8,099,871 – 8,099,979 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -39.34 |
| Consensus MFE | -33.78 |
| Energy contribution | -34.02 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8099871 108 + 27905053 -GGCGCUGCGGUUGCCGAGGCAGCCGCC---GCCAAGAAAAAUAGCCAAGCUUGGAAAAUGCGUUGGAGUAAUAGCCGGUAACG----AACCCGAGUUGGUGGAUGGA----UUUCCAUA -((((..((((((((....)))))))))---)))..........(((((.(((((......(((((..(.......)..)))))----...))))))))))..((((.----...)))). ( -40.90) >DroSec_CAF1 51516 112 + 1 -AGCGCUGCGGUUGCCGGGGCAGCCGCC---GCCAAGAAAAAUAGCCAAGCUUGGAAAAUGCGUUGGAGUAAUAGUCGGUAACGAACGAACCCGAGCUGGUGGAUGGA----UUUCCAUA -.(.((.((((((((....)))))))).---)))..........((((.((((((....(((......)))....(((....)))......))))))))))..((((.----...)))). ( -41.00) >DroSim_CAF1 23587 112 + 1 -GGAGCUGCGGUUGCCGAGGCAGCCGCC---GCCAAGAAAAAUAGUCAAGCUUGGAAAAUGCGUUGGAGUAAUAGUCGGUAACGAACGAACCCGAGCUGGUGGAUGGA----UUUCCAUA -.(.((.((((((((....)))))))).---)))..............(((((((....(((......)))....(((....)))......))))))).(((((....----..))))). ( -38.60) >DroEre_CAF1 26602 112 + 1 GGACUCUGCGGUUACCGAGGCAGCCGCCGUCGCCAAGAAAAAUGGCCAAGUUUGGAAAAUGCGUUGGAGUAAUAGUCGGUAACG----AGCCCGAGCUUGUGGAUGGA----UUUCCAUA (((....((((((.(....).))))))((((((((.......))))(((((((((....(((......)))....(((....))----)..)))))))))..))))..----..)))... ( -35.30) >DroYak_CAF1 19943 112 + 1 -GGCUCUGCGGUUGCCGAGGCAGCCGCC---GCCAAGAAAAAUAGCCAAGUUUGGAAAAUGCGUUGGAGUAAUAGUCGGUAACG----AACCCGAGCUUGUGGAUGGAUGCAUUUCCAUA -(((...((((((((....)))))))).---)))...........((((((((((....(((......)))....(((....))----)..))))))))).).(((((......))))). ( -40.90) >consensus _GGCGCUGCGGUUGCCGAGGCAGCCGCC___GCCAAGAAAAAUAGCCAAGCUUGGAAAAUGCGUUGGAGUAAUAGUCGGUAACG____AACCCGAGCUGGUGGAUGGA____UUUCCAUA .(((...((((((((....))))))))....)))...........((((((((((......(((((..(.......)..))))).......)))))))..)))(((((......))))). (-33.78 = -34.02 + 0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:01 2006