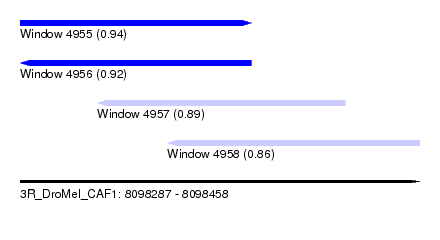
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,098,287 – 8,098,458 |
| Length | 171 |
| Max. P | 0.938210 |

| Location | 8,098,287 – 8,098,386 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.03 |
| Mean single sequence MFE | -33.36 |
| Consensus MFE | -20.75 |
| Energy contribution | -20.75 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8098287 99 + 27905053 UAUAAUUAACCACAUGUGUGUGUGUGUGUGGGG--------------GUUUGUCCGGGGAAGCGGUUUAGCAUGUCC-------UUUGACUGCUUCCGCUCGUCGGUUAAAGUUACUUCC .........(((((..(......)..)))))((--------------.((((.((((.((.((((...((((.(((.-------...))))))).)))))).)))).)))).))...... ( -33.80) >DroSec_CAF1 49949 97 + 1 UAUAAUUAACCACAUGUGUGU--GUGUGUGGGG--------------GUUUGUCGGGGGAAGCGGUUUAGCAUGUCC-------UUUGACUGCUUCCGCUCGAUGGUUAAAGUUACUUCC .....((((((((((....))--))(.((((((--------------((..(((((((((.((......))...)))-------)))))).)))))))).)...)))))).......... ( -34.10) >DroSim_CAF1 22033 93 + 1 UAUAAUUAACCACAUGUGU------GUGUGGGG--------------GUUUGUCAGGGGAAGCGGUUUAGCAUGUCC-------UUUGACUGCUUCCGCUCGAUGGUUAAAGUUACUUCC .....(((((((......(------(.((((((--------------((..(((((((((.((......))...)))-------)))))).)))))))).)).))))))).......... ( -33.30) >DroEre_CAF1 25007 89 + 1 UAUAAUUAACCACAUGUGUGUGU------------------------------C-GGGGAAGCGGUUUAGCAUGUCCUUUGUCCUUUGACUGCUUCCGCCCGUUGGUUAAAGUUACUUCC ..((((((((((((....)))).------------------------------.-((((((((((((..(((.......))).....)))))))))).)).))))))))........... ( -26.00) >DroYak_CAF1 18425 113 + 1 UAUAAUUAACCACAUGUGUAUGUGUGUGUGGGGGGGGGGGGGGGGGGUUUUGUCGGGGGAAGCGGUUUAGCAUGUCC-------UUUGACUGCUUCCGCUCGUCGGUUAAAGUUACUUCC ..(((((..(((((..(......)..)))))....((.(((.((((((...(((((((((.((......))...)))-------)))))).)))))).))).)))))))........... ( -39.60) >consensus UAUAAUUAACCACAUGUGUGUGUGUGUGUGGGG______________GUUUGUCGGGGGAAGCGGUUUAGCAUGUCC_______UUUGACUGCUUCCGCUCGUUGGUUAAAGUUACUUCC .....((((((((....))....................................(.((((((((((....................)))))))))).).....)))))).......... (-20.75 = -20.75 + 0.00)

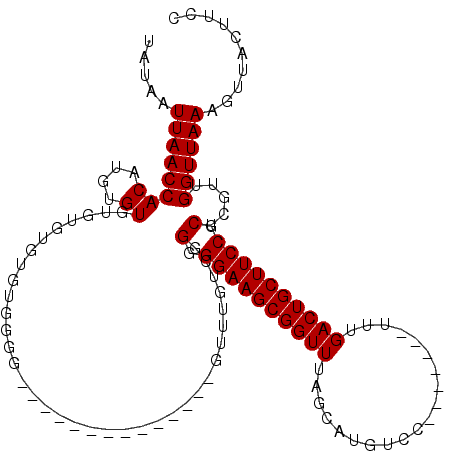
| Location | 8,098,287 – 8,098,386 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.03 |
| Mean single sequence MFE | -22.04 |
| Consensus MFE | -19.12 |
| Energy contribution | -18.96 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8098287 99 - 27905053 GGAAGUAACUUUAACCGACGAGCGGAAGCAGUCAAA-------GGACAUGCUAAACCGCUUCCCCGGACAAAC--------------CCCCACACACACACACACAUGUGGUUAAUUAUA .....((((.....(((..((((((.(((((((...-------.))).))))...))))))...)))......--------------.........((((......))))))))...... ( -24.10) >DroSec_CAF1 49949 97 - 1 GGAAGUAACUUUAACCAUCGAGCGGAAGCAGUCAAA-------GGACAUGCUAAACCGCUUCCCCCGACAAAC--------------CCCCACACAC--ACACACAUGUGGUUAAUUAUA ..........((((((...((((((.(((((((...-------.))).))))...))))))............--------------........((--(......)))))))))..... ( -20.80) >DroSim_CAF1 22033 93 - 1 GGAAGUAACUUUAACCAUCGAGCGGAAGCAGUCAAA-------GGACAUGCUAAACCGCUUCCCCUGACAAAC--------------CCCCACAC------ACACAUGUGGUUAAUUAUA ..........((((((((...((....)).((((..-------(((...((......)).)))..))))....--------------........------......))))))))..... ( -21.70) >DroEre_CAF1 25007 89 - 1 GGAAGUAACUUUAACCAACGGGCGGAAGCAGUCAAAGGACAAAGGACAUGCUAAACCGCUUCCCC-G------------------------------ACACACACAUGUGGUUAAUUAUA ..........((((((....((.((((((.(((....)))...((..........))))))))))-.------------------------------(((......)))))))))..... ( -21.70) >DroYak_CAF1 18425 113 - 1 GGAAGUAACUUUAACCGACGAGCGGAAGCAGUCAAA-------GGACAUGCUAAACCGCUUCCCCCGACAAAACCCCCCCCCCCCCCCCCCACACACACAUACACAUGUGGUUAAUUAUA ..........((((((...((((((.(((((((...-------.))).))))...))))))....................................((((....))))))))))..... ( -21.90) >consensus GGAAGUAACUUUAACCAACGAGCGGAAGCAGUCAAA_______GGACAUGCUAAACCGCUUCCCCCGACAAAC______________CCCCACACACACACACACAUGUGGUUAAUUAUA ..........((((((...((((((.(((((((...........))).))))...))))))........................................((....))))))))..... (-19.12 = -18.96 + -0.16)

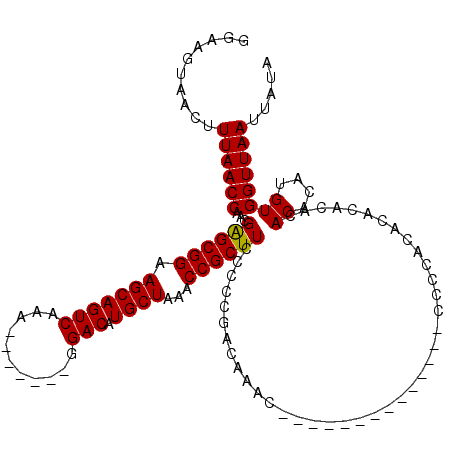

| Location | 8,098,320 – 8,098,426 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.07 |
| Mean single sequence MFE | -28.54 |
| Consensus MFE | -25.40 |
| Energy contribution | -25.56 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8098320 106 - 27905053 UCAGAACGCUUACAGAGUUACACACGUGACACUAACGGGAGGAAGUAACUUUAACCGACGAGCGGAAGCAGUCAAA-------GGACAUGCUAAACCGCUUCCCCGGACAAAC------- .............((((((((.(.(((.......)))...)...))))))))..(((..((((((.(((((((...-------.))).))))...))))))...)))......------- ( -27.70) >DroSec_CAF1 49980 106 - 1 UCAGAUCGCUUACAGAGUUACACACGUGACACUAACGGGAGGAAGUAACUUUAACCAUCGAGCGGAAGCAGUCAAA-------GGACAUGCUAAACCGCUUCCCCCGACAAAC------- .............((.(((((....))))).))..((((.((((((........((.......)).(((((((...-------.))).)))).....))))))))))......------- ( -29.30) >DroSim_CAF1 22060 106 - 1 UCAGAUCGCUUACAGAGUUACACACGUGACACUAACGGGAGGAAGUAACUUUAACCAUCGAGCGGAAGCAGUCAAA-------GGACAUGCUAAACCGCUUCCCCUGACAAAC------- ((((.........((.(((((....))))).))...((((((............))....(((((.(((((((...-------.))).))))...))))))))))))).....------- ( -27.90) >DroEre_CAF1 25030 106 - 1 UUAGAUCGCUUACGGAGUUACACACGUGACACUAACGGGAGGAAGUAACUUUAACCAACGGGCGGAAGCAGUCAAAGGACAAAGGACAUGCUAAACCGCUUCCCC-G------------- .............((.(((((....)))))(((..(....)..)))........))....((.((((((.(((....)))...((..........))))))))))-.------------- ( -25.70) >DroYak_CAF1 18465 113 - 1 UUAGAUCGCUUACAGAGUUACACACGUGACACUAACGGGAGGAAGUAACUUUAACCGACGAGCGGAAGCAGUCAAA-------GGACAUGCUAAACCGCUUCCCCCGACAAAACCCCCCC ((((.((.......))(((((....))))).))))((((.((((((........(((.....))).(((((((...-------.))).)))).....))))))))))............. ( -32.10) >consensus UCAGAUCGCUUACAGAGUUACACACGUGACACUAACGGGAGGAAGUAACUUUAACCAACGAGCGGAAGCAGUCAAA_______GGACAUGCUAAACCGCUUCCCCCGACAAAC_______ .............((.(((((....))))).))..((((.((((((........((.......)).(((((((...........))).)))).....))))))))))............. (-25.40 = -25.56 + 0.16)

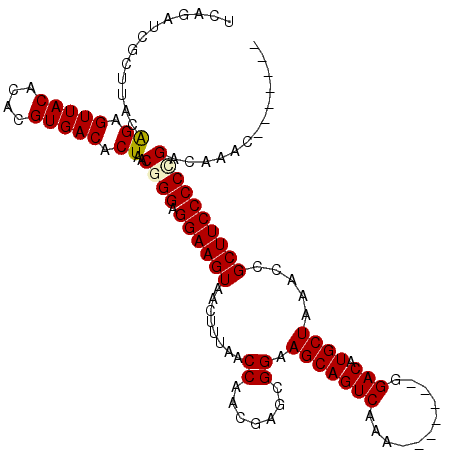
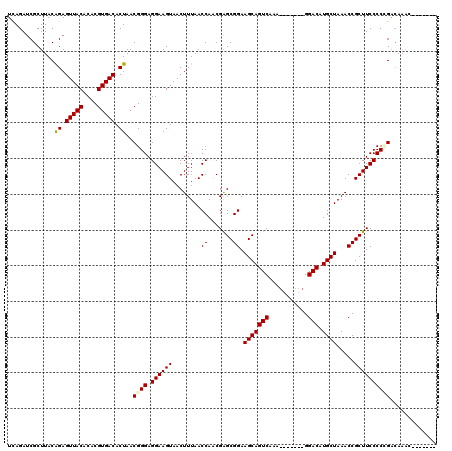
| Location | 8,098,350 – 8,098,458 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.15 |
| Mean single sequence MFE | -36.46 |
| Consensus MFE | -27.97 |
| Energy contribution | -27.93 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8098350 108 - 27905053 UAUGCCCUCGGGGCAAAGGAAGGGG--------ACUUCCCUCAGAACGCUUACAGAGUUACACACGUGACACUAACGGGAGGAAGUAACUUUAACCGACGAGCGGAAGCAGUCAAA---- ..(((((...)))))..((((((..--------(((((((((((....))...((.(((((....))))).))...))).))))))..))))..))(((..((....)).)))...---- ( -35.90) >DroSec_CAF1 50010 116 - 1 UAUGCCCUCGGGGCAAAGGAAGGGGACUGAGGGACUUCCCUCAGAUCGCUUACAGAGUUACACACGUGACACUAACGGGAGGAAGUAACUUUAACCAUCGAGCGGAAGCAGUCAAA---- ..(((((...)))))..((.(((.(((((((((....))))))).)).)))..((((((((.(.(((.......)))...)...))))))))..)).....((....)).......---- ( -39.60) >DroSim_CAF1 22090 116 - 1 UAUGCCCUCGGGGCAAAGGAAGGGGACUGAGGGACUUCCCUCAGAUCGCUUACAGAGUUACACACGUGACACUAACGGGAGGAAGUAACUUUAACCAUCGAGCGGAAGCAGUCAAA---- ..(((((...)))))..((.(((.(((((((((....))))))).)).)))..((((((((.(.(((.......)))...)...))))))))..)).....((....)).......---- ( -39.60) >DroEre_CAF1 25056 112 - 1 UAUGCCCUCGGGGCAAGGAAAGGGG--------ACUUCCCUUAGAUCGCUUACGGAGUUACACACGUGACACUAACGGGAGGAAGUAACUUUAACCAACGGGCGGAAGCAGUCAAAGGAC ..(((((...))))).((((((..(--------.((((((((((.(((....))).(((((....))))).)))).))))))...)..))))..)).....((....)).(((....))) ( -32.50) >DroYak_CAF1 18502 108 - 1 UAUGCCCUCGGGGCAAAGGAAGGGG--------ACUUCCCUUAGAUCGCUUACAGAGUUACACACGUGACACUAACGGGAGGAAGUAACUUUAACCGACGAGCGGAAGCAGUCAAA---- ..(((((...)))))..((((((..--------(((((((((...((.......))(((((....)))))......))).))))))..))))..))(((..((....)).)))...---- ( -34.70) >consensus UAUGCCCUCGGGGCAAAGGAAGGGG________ACUUCCCUCAGAUCGCUUACAGAGUUACACACGUGACACUAACGGGAGGAAGUAACUUUAACCAACGAGCGGAAGCAGUCAAA____ ..(((((...))))).......((.........(((((((((...((.......))(((((....)))))......))).))))))........)).....((....))........... (-27.97 = -27.93 + -0.04)

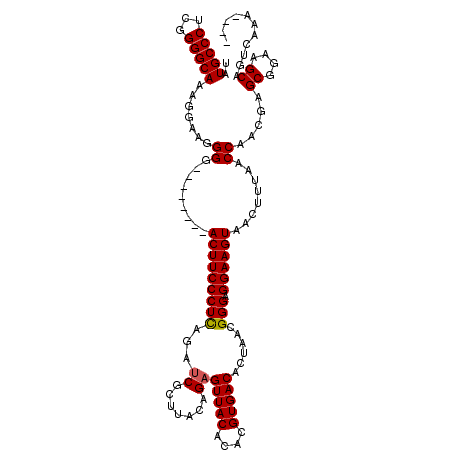
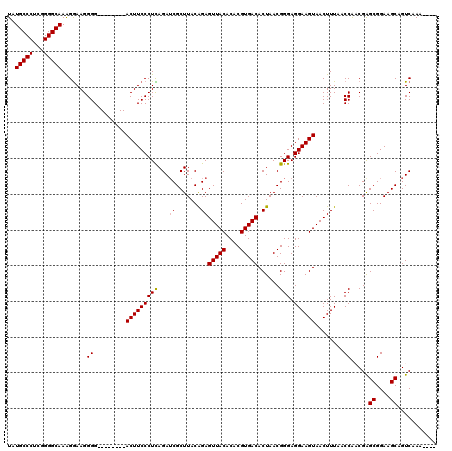
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:56 2006