
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,097,559 – 8,097,792 |
| Length | 233 |
| Max. P | 0.972622 |

| Location | 8,097,559 – 8,097,672 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.34 |
| Mean single sequence MFE | -22.26 |
| Consensus MFE | -19.42 |
| Energy contribution | -19.02 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
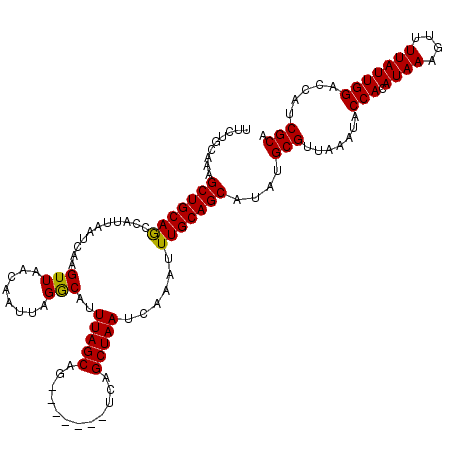
>3R_DroMel_CAF1 8097559 113 + 27905053 UUCUGCAAUGCUGCAGCCAUUAAUCAAGUUAACAAUUAGGCAUUUAGCAG-------UCAGCUAAUCAAAUUUGCAGCAUAUGCGUUAAAUACCACAUAAAGUUUUAUUGGACCAUCGCA ....((((((((((((((....................)))..(((((..-------...))))).......)))))))).)))........(((.((((....)))))))......... ( -25.35) >DroSec_CAF1 49221 113 + 1 UUCUGCAAAGCUGCAGCCAUUAAUCAAGUUAACAAUUAGACAUUUAGCAG-------UCAGCUAAUCAAAUUUGCAGCAUAUGCGUUAAACGCCACAUAAAGUUUUAUUGGACCAUCGCA .........(((((((...........(((........)))..(((((..-------...)))))......)))))))...((((.......(((.((((....))))))).....)))) ( -21.70) >DroSim_CAF1 21310 113 + 1 UUCUGCAAAGCUGCAGCCAUUAAUCAAGUUAACAAUUAGGCAUUUAGCAG-------UCAGCUAAUCAAAUUUGCAGCAUAUGCGUUAAAUACCACAUAAAGUUUUAUUGGACCAUCGCA ....((...(((((((((....................)))..(((((..-------...))))).......))))))..(((.(((.((((..((.....))..)))).)))))).)). ( -23.35) >DroEre_CAF1 24302 113 + 1 UUCUGCAAAGCUGCAACCAUUAAUCAAGUUAACAAUUAGGCAUUUAGCAG-------UCAGCUAAUCAAAUUUGCAGCAUAUGCGUUAAAUACCACAUAAAGUUUUAUUGGACCAUCGCA ....((...(((((((...........(((........)))..(((((..-------...)))))......)))))))..(((.(((.((((..((.....))..)))).)))))).)). ( -20.90) >DroYak_CAF1 17669 120 + 1 UUCUGCAAAGCUGCAACCAUUAAUCAAGUUAACAAUUAGGCAUUUAGCAGUCAGCAGUCAGCUAAUCAAAUUUGCAGCAUAUGCGUUAAAUACCACAUAAAGUUUUAUUGGACCAUCGCC ....((...(((((((......................(((........)))(((.....)))........)))))))..(((.(((.((((..((.....))..)))).)))))).)). ( -20.00) >consensus UUCUGCAAAGCUGCAGCCAUUAAUCAAGUUAACAAUUAGGCAUUUAGCAG_______UCAGCUAAUCAAAUUUGCAGCAUAUGCGUUAAAUACCACAUAAAGUUUUAUUGGACCAUCGCA .........(((((((...........(((........)))..(((((............)))))......)))))))....(((.......(((.((((....))))))).....))). (-19.42 = -19.02 + -0.40)

| Location | 8,097,559 – 8,097,672 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.34 |
| Mean single sequence MFE | -28.15 |
| Consensus MFE | -26.40 |
| Energy contribution | -26.00 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
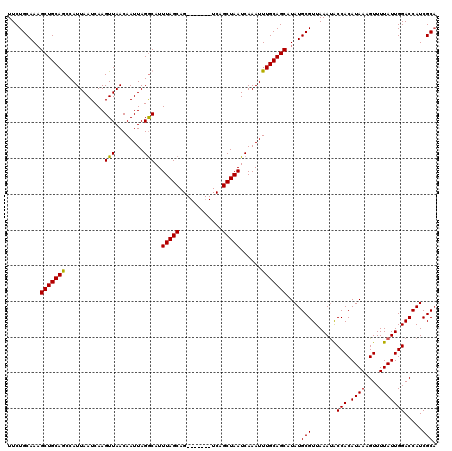
>3R_DroMel_CAF1 8097559 113 - 27905053 UGCGAUGGUCCAAUAAAACUUUAUGUGGUAUUUAACGCAUAUGCUGCAAAUUUGAUUAGCUGA-------CUGCUAAAUGCCUAAUUGUUAACUUGAUUAAUGGCUGCAGCAUUGCAGAA ((((((...............((((((........)))))).((((((.......(((((...-------..)))))..(((((((((......))))))..)))))))))))))))... ( -30.90) >DroSec_CAF1 49221 113 - 1 UGCGAUGGUCCAAUAAAACUUUAUGUGGCGUUUAACGCAUAUGCUGCAAAUUUGAUUAGCUGA-------CUGCUAAAUGUCUAAUUGUUAACUUGAUUAAUGGCUGCAGCUUUGCAGAA (((((................((((((........)))))).((((((.......(((((...-------..)))))..(((((((((......))))))..))))))))).)))))... ( -26.40) >DroSim_CAF1 21310 113 - 1 UGCGAUGGUCCAAUAAAACUUUAUGUGGUAUUUAACGCAUAUGCUGCAAAUUUGAUUAGCUGA-------CUGCUAAAUGCCUAAUUGUUAACUUGAUUAAUGGCUGCAGCUUUGCAGAA (((((................((((((........)))))).((((((.......(((((...-------..)))))..(((((((((......))))))..))))))))).)))))... ( -29.10) >DroEre_CAF1 24302 113 - 1 UGCGAUGGUCCAAUAAAACUUUAUGUGGUAUUUAACGCAUAUGCUGCAAAUUUGAUUAGCUGA-------CUGCUAAAUGCCUAAUUGUUAACUUGAUUAAUGGUUGCAGCUUUGCAGAA (((((................((((((........)))))).(((((((..((((((((.(((-------(.((.....))......)))).).)))))))...))))))).)))))... ( -27.10) >DroYak_CAF1 17669 120 - 1 GGCGAUGGUCCAAUAAAACUUUAUGUGGUAUUUAACGCAUAUGCUGCAAAUUUGAUUAGCUGACUGCUGACUGCUAAAUGCCUAAUUGUUAACUUGAUUAAUGGUUGCAGCUUUGCAGAA .((((................((((((........)))))).(((((((..((((((((.((((........((.....))......)))).).)))))))...))))))).)))).... ( -27.24) >consensus UGCGAUGGUCCAAUAAAACUUUAUGUGGUAUUUAACGCAUAUGCUGCAAAUUUGAUUAGCUGA_______CUGCUAAAUGCCUAAUUGUUAACUUGAUUAAUGGCUGCAGCUUUGCAGAA .((((................((((((........)))))).((((((.......(((((............)))))..(((((((((......))))))..))))))))).)))).... (-26.40 = -26.00 + -0.40)



| Location | 8,097,632 – 8,097,752 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -29.48 |
| Energy contribution | -29.04 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
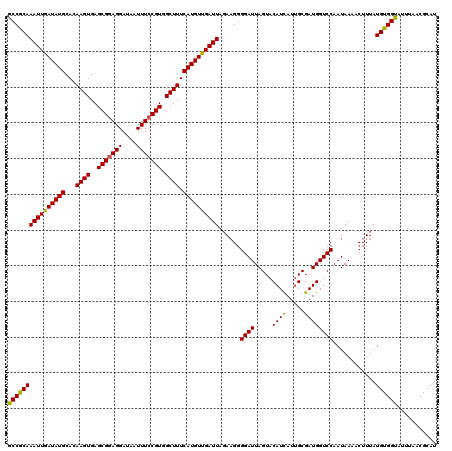
>3R_DroMel_CAF1 8097632 120 - 27905053 GCCGCAAAUUGAUAUGCACAAGUGAGCGGAGGAUAAUUUCCGUGGCUUUCAUGUUGAUUAGAAGGGGAUUAGUACAUCAUUGCGAUGGUCCAAUAAAACUUUAUGUGGUAUUUAACGCAU (((((((((..(((((...((((..(((((((....))))))).)))).)))))..)))......((((((...((....))...))))))............))))))........... ( -31.00) >DroSec_CAF1 49294 120 - 1 GCCGCAAAUUGAUAUGCACAAGUGAGCGGAGGAUAAUUUCCGUGGCUUUCAUGUUGAUUAGAAGGGGAUUAGUACAUUAUUGCGAUGGUCCAAUAAAACUUUAUGUGGCGUUUAACGCAU (((((((((..(((((...((((..(((((((....))))))).)))).)))))..)))......((((((...((....))...))))))............))))))........... ( -33.40) >DroSim_CAF1 21383 120 - 1 GCCGCAAAUUGAUAUGCACAAGUGAGCGGAGGAUAAUUUCCGUGGCUUUCAUGUUGAUUACAAGGGGAUUAGUACAUCAUUGCGAUGGUCCAAUAAAACUUUAUGUGGUAUUUAACGCAU (((((((((..(((((...((((..(((((((....))))))).)))).)))))..)))......((((((...((....))...))))))............))))))........... ( -31.00) >DroEre_CAF1 24375 120 - 1 GCCGCAAAUUGAUAUGCACAAGUGAGCGGAGGAUAAUUUCCGUGGCUUUCAUGUUGAUUAGAAGGGGAUUAGUACAUCAUUGCGAUGGUCCAAUAAAACUUUAUGUGGUAUUUAACGCAU (((((((((..(((((...((((..(((((((....))))))).)))).)))))..)))......((((((...((....))...))))))............))))))........... ( -31.00) >DroYak_CAF1 17749 120 - 1 GCCACAAAUUAAUAUGCACAAGUGAGCGAAGGAUAAUUUGCGUGGCUUUCAUGUUGAUUAGACGGGGAUUAGUACAUCAUGGCGAUGGUCCAAUAAAACUUUAUGUGGUAUUUAACGCAU ((((((((((((((((...((((.((((((......))))).).)))).))))))))))......((((.....((((.....))))))))............))))))........... ( -29.00) >consensus GCCGCAAAUUGAUAUGCACAAGUGAGCGGAGGAUAAUUUCCGUGGCUUUCAUGUUGAUUAGAAGGGGAUUAGUACAUCAUUGCGAUGGUCCAAUAAAACUUUAUGUGGUAUUUAACGCAU ((((((((((((((((...((((..(((((((....))))))).)))).))))))))))......((((.....((((.....))))))))............))))))........... (-29.48 = -29.04 + -0.44)


| Location | 8,097,672 – 8,097,792 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -34.36 |
| Consensus MFE | -30.62 |
| Energy contribution | -30.54 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
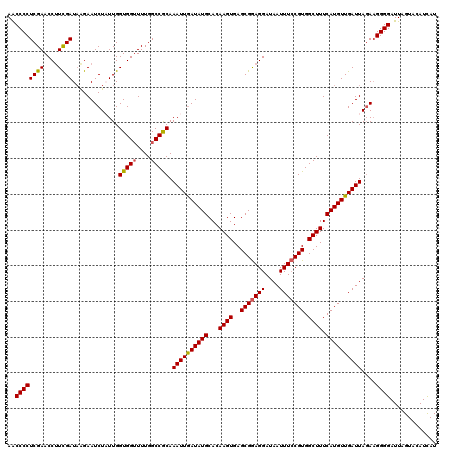
>3R_DroMel_CAF1 8097672 120 - 27905053 AACCCCUCGAACCUUUGAUUGGAAUCUAUUGGUGGUUUUGGCCGCAAAUUGAUAUGCACAAGUGAGCGGAGGAUAAUUUCCGUGGCUUUCAUGUUGAUUAGAAGGGGAUUAGUACAUCAU ..(((((....((.......))..(((....(((((....))))).(((..(((((...((((..(((((((....))))))).)))).)))))..)))))))))))............. ( -33.90) >DroSec_CAF1 49334 120 - 1 AACCCCUCGAACCUUCGAUAAGAAUCUUUUGGUGGUUUUGGCCGCAAAUUGAUAUGCACAAGUGAGCGGAGGAUAAUUUCCGUGGCUUUCAUGUUGAUUAGAAGGGGAUUAGUACAUUAU ..((((((((....))))......(((....(((((....))))).(((..(((((...((((..(((((((....))))))).)))).)))))..)))))).))))............. ( -33.80) >DroSim_CAF1 21423 120 - 1 AACCCCUCGAACCUUCGAUAAGAAUCUUUUGGUGGUUUUGGCCGCAAAUUGAUAUGCACAAGUGAGCGGAGGAUAAUUUCCGUGGCUUUCAUGUUGAUUACAAGGGGAUUAGUACAUCAU ..((((((((....))))..........((((((((....))))).(((..(((((...((((..(((((((....))))))).)))).)))))..))).)))))))............. ( -34.20) >DroEre_CAF1 24415 120 - 1 AGCCCCUCGAACCAUCGAUAAGAAUCUAUUGGUGGGUUUGGCCGCAAAUUGAUAUGCACAAGUGAGCGGAGGAUAAUUUCCGUGGCUUUCAUGUUGAUUAGAAGGGGAUUAGUACAUCAU ..((((((((((((((((((......))))))).))))))......(((..(((((...((((..(((((((....))))))).)))).)))))..)))...)))))............. ( -37.20) >DroYak_CAF1 17789 120 - 1 AACCCCUCGAACCUUCGAUAAGAAUCUAUUGGUGGUUUUGGCCACAAAUUAAUAUGCACAAGUGAGCGAAGGAUAAUUUGCGUGGCUUUCAUGUUGAUUAGACGGGGAUUAGUACAUCAU ..((((((((....))))......(((....(((((....))))).((((((((((...((((.((((((......))))).).)))).))))))))))))).))))............. ( -32.70) >consensus AACCCCUCGAACCUUCGAUAAGAAUCUAUUGGUGGUUUUGGCCGCAAAUUGAUAUGCACAAGUGAGCGGAGGAUAAUUUCCGUGGCUUUCAUGUUGAUUAGAAGGGGAUUAGUACAUCAU ..((((((((....)))).............(((((....))))).((((((((((...((((..(((((((....))))))).)))).))))))))))....))))............. (-30.62 = -30.54 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:50 2006