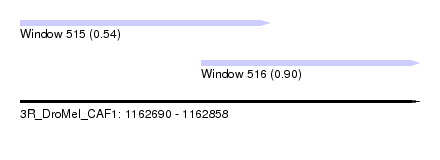
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,162,690 – 1,162,858 |
| Length | 168 |
| Max. P | 0.898028 |

| Location | 1,162,690 – 1,162,795 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 95.24 |
| Mean single sequence MFE | -23.75 |
| Consensus MFE | -19.42 |
| Energy contribution | -20.43 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
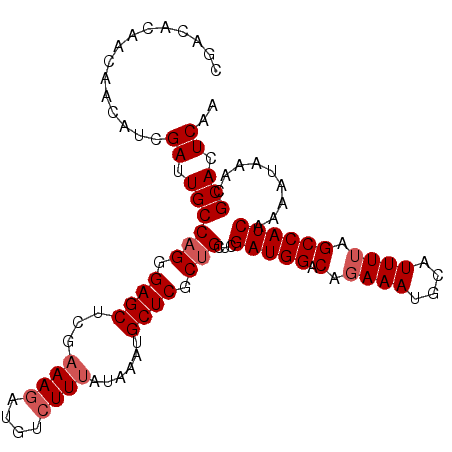
>3R_DroMel_CAF1 1162690 105 + 27905053 CGACACAACAACAUCGAUUGCCAGGGAGCUCAAAAUAUGUCUUCAUAAAUGCUCGCCGCUCGAUGGACAGAAAUGCAUUUUAGCCAUCAAAAUAAACGCACUCAA ...............((.(((..((((((......((((....))))...)))).))....(((((..((((.....))))..))))).........))).)).. ( -21.50) >DroSec_CAF1 98335 105 + 1 CGACACAACAACAUCGAUUGCCAGGGAGCUCGAAAGAUGUCUUUAUAAAUGCUCGCUGCUCGAUGGACAGAAAUGCAUUUUAGCCAUCAAAAUAAACGCACUCAA ...............((.((((((.((((...((((....))))......)))).)))...(((((..((((.....))))..))))).........))).)).. ( -24.50) >DroSim_CAF1 94616 105 + 1 CGACACAACAACAUCGAUUGCCAGGGAGCUCGAAAGAUGUCUUUAUAAAUGCUCGCUGCUCGAUGGACAGAAAUGCAUUUUAGCCAUCAAAAUAAACGCACUCAA ...............((.((((((.((((...((((....))))......)))).)))...(((((..((((.....))))..))))).........))).)).. ( -24.50) >DroEre_CAF1 96194 105 + 1 CGACACAACAACAUCGAUUGCCAGAGAGCUCGAAAGAUGUCUUUAUAAAUGCUCACUGCUUGAUGGACAGAAAUGCAUUUAAGCCAUCGAUAUAAACGCACUCAA ............((((((.((.(((((..((....))..))))).(((((((...(((((....)).)))....))))))).)).)))))).............. ( -24.50) >consensus CGACACAACAACAUCGAUUGCCAGGGAGCUCGAAAGAUGUCUUUAUAAAUGCUCGCUGCUCGAUGGACAGAAAUGCAUUUUAGCCAUCAAAAUAAACGCACUCAA ...............((.((((((.((((...((((....))))......)))).)))...(((((.(.((((....)))).)))))).........))).)).. (-19.42 = -20.43 + 1.00)

| Location | 1,162,766 – 1,162,858 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 77.69 |
| Mean single sequence MFE | -17.10 |
| Consensus MFE | -6.96 |
| Energy contribution | -7.08 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
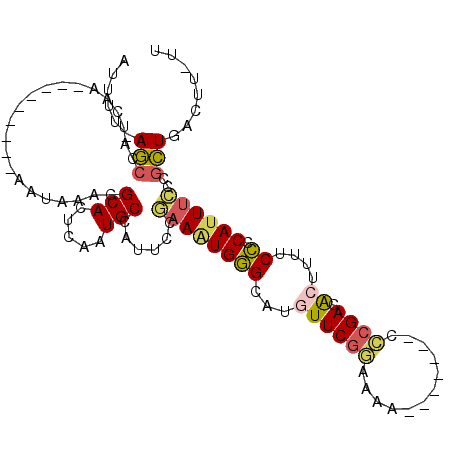
>3R_DroMel_CAF1 1162766 92 + 27905053 AUUUU-AGCCAUCAA---------AAUAAACGCACUCAAUGCCAUUCCGAAGUGGGAAUGUUCGGAACA-------CCCGACACUUUUCCCCAUUUCCCGCUAACUU-UU ...((-(((......---------.......(((.....)))......((((((((((.((((((....-------.)))).))..)).))))))))..)))))...-.. ( -21.40) >DroSec_CAF1 98411 92 + 1 AUUUU-AGCCAUCAA---------AAUAAACGCACUCAAUGCCAUUCCGAAAUGGGCAUGUUCGGAAAA-------CCCGACACUUUUCCCCAUUUCCCGCUGACUU-UU ...((-(((......---------.......(((.....)))......((((((((...((((((....-------.)))).)).....))))))))..)))))...-.. ( -21.40) >DroSim_CAF1 94692 92 + 1 AUUUU-AGCCAUCAA---------AAUAAACGCACUCAAUGCCAUUCCGAAAUGGGCAUGUUCGGAAAA-------CCCGACACUUUUCCCCAUUUCCCGCUGACUU-UU ...((-(((......---------.......(((.....)))......((((((((...((((((....-------.)))).)).....))))))))..)))))...-.. ( -21.40) >DroEre_CAF1 96270 92 + 1 AUUUA-AGCCAUCGA---------UAUAAACGCACUCAAUGCCAUCCCAAAAUGGGCUUGUUCGAAAAA-------CCCGACGAU-UUCCACAUUCCCCACUCACUUUUU ...((-((((.....---------.......(((.....)))(((......))))))))).(((.....-------..)))....-........................ ( -9.40) >DroAna_CAF1 92558 110 + 1 AUUUUGAACCAUCAAAUUCCGUAAAAUAAACGCACUCAAUGCCAUUCAAAAUUGUGCAUGUUCAAAAAAAAAUCCUUUCGAUGAUUUCCACCACUUUUCGUUUACAUUGC .((((((((..........(((.......)))......((((((........)).))))))))))))............(((((.............)))))........ ( -11.92) >consensus AUUUU_AGCCAUCAA_________AAUAAACGCACUCAAUGCCAUUCCGAAAUGGGCAUGUUCGGAAAA_______CCCGACACUUUUCCCCAUUUCCCGCUGACUU_UU ......(((......................(((.....)))......((((((((...((((((............)))).))....)).))))))..)))........ ( -6.96 = -7.08 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:57 2006