
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,091,255 – 8,091,633 |
| Length | 378 |
| Max. P | 0.992315 |

| Location | 8,091,255 – 8,091,360 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.35 |
| Mean single sequence MFE | -34.12 |
| Consensus MFE | -28.42 |
| Energy contribution | -28.42 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8091255 105 + 27905053 CUUGGUCAGUCAUGCACUCAAUCAGUGGCAGGACACUCCGGCCAGGACAACAGGCGCAGUUCACCCCCUGCG-------ACAUGUAA-AGGGUACUCGCCCCCACCCUCGCC-------U ...((...(((.((((((.....))).))).)))...))(((.(((......((((.(((..((((..(((.-------....))).-.))))))))))).....))).)))-------. ( -33.40) >DroSec_CAF1 43027 113 + 1 CUUGGUCAGUCAUGCACUCAAUCAGUGGCAGGACACUGCGGCCAGGACAACAGGCGCAGUUCACCCCCUGCG-------ACAUGUAAAAGGGUACUCGCCCUCACCCUCGCCCUCAUCCA (((((((.......((((.....))))((((....)))))))))))......(((((((........)))).-------.........(((((....))))).......)))........ ( -35.00) >DroSim_CAF1 15032 113 + 1 CUUGGUCAGUCAUGCACUCAAUCAGUGGCAGGACACUCCGGCCAGGACAACAGGCGCAGUUCACCCCCUGCG-------ACAUGUAAAAGGGUACUCGCCCUCACCCUCGCCCUCACCCA ...((...(((.((((((.....))).))).)))...))(((.(((...(((..(((((........)))))-------...)))...(((((....)))))...))).)))........ ( -33.90) >DroEre_CAF1 18069 113 + 1 CUUGGUCAGUCAUGCACUCAAUCAGUGGCAGGACACUCCGGCCAGGACAACAGGCGCAGUUCACCCCCUGCUACCCUUCACAUGUAA-AGGGUACUCGCCCUCGCCAUC------GCCCU (((((((.(((((...........))))).((.....)))))))))......(((((((........))))(((((((........)-)))))).........)))...------..... ( -34.20) >consensus CUUGGUCAGUCAUGCACUCAAUCAGUGGCAGGACACUCCGGCCAGGACAACAGGCGCAGUUCACCCCCUGCG_______ACAUGUAA_AGGGUACUCGCCCUCACCCUCGCC___ACCCA (((((((.(((.((((((.....))).))).))).....)))))))......((((.(((..((((..(((............)))...))))))))))).................... (-28.42 = -28.42 + 0.00)

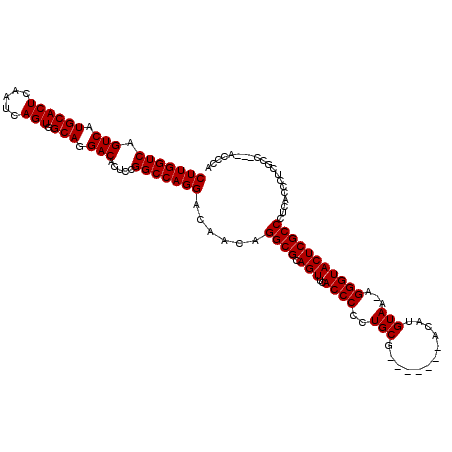

| Location | 8,091,328 – 8,091,440 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.60 |
| Mean single sequence MFE | -30.94 |
| Consensus MFE | -27.46 |
| Energy contribution | -27.98 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8091328 112 + 27905053 CAUGUAA-AGGGUACUCGCCCCCACCCUCGCC-------UCUCCCUCCUUUAUGAAGUGCAGUUUUUAGUUCCGUUUCCCUCCGCCAGGAAGUCAGACUUCCUAAGCGGAACUCGCUGCA ((((...-((((.....((..........)).-------...))))....))))...((((((....((((((((((.........((((((.....)))))))))))))))).)))))) ( -32.00) >DroSec_CAF1 43100 120 + 1 CAUGUAAAAGGGUACUCGCCCUCACCCUCGCCCUCAUCCACUCCAUCCUUCGUGAAGUGCAGUUUUUAGUUCUGUUUCCCCCCGCCAGGAAGUCAGACUUCCAAAGCGGAACUCGCUGCA ........(((((....))))).................(((.(((.....))).)))(((((....((((((((((..........(((((.....))))).)))))))))).))))). ( -28.40) >DroSim_CAF1 15105 120 + 1 CAUGUAAAAGGGUACUCGCCCUCACCCUCGCCCUCACCCACUCCAUCCUUCGUGAAGUGCAGUUUUUAGUUCCUUUUCUCCCCGCCAGGAAGUCAGACUUCCAAAGCGGAACUCGCUGCA ........(((((....))))).................(((.(((.....))).)))(((((....((((((..........((..(((((.....)))))...)))))))).))))). ( -26.70) >DroEre_CAF1 18149 113 + 1 CAUGUAA-AGGGUACUCGCCCUCGCCAUC------GCCCUCGCCCUCCUUCAUGGACUGCAGUUUGUAGUUCAGUUUCCCCCCGCCAGGAAGUCGGACUUCCUAAGCGGAACUCGCUGCA ..((((.-(((((....((....))....------))))).((.........((((((((.....))))))))........((((.((((((.....))))))..)))).....)))))) ( -33.40) >DroYak_CAF1 11495 118 + 1 CAUGUAA-AGGGUACUCGCCCUCACCCUCACCCUCACCCUCGUCCUCCUUCAUGAAGUGCAGUUUGUAGUUCCGCUUCC-CUCGCCAGGAAGUCAGACUUCCUAAGCGGAACUCGCUGCA .......-(((((....))))).................((((........))))..((((((..(.((((((((((..-......((((((.....))))))))))))))))))))))) ( -34.20) >consensus CAUGUAA_AGGGUACUCGCCCUCACCCUCGCCCUCACCCUCUCCCUCCUUCAUGAAGUGCAGUUUUUAGUUCCGUUUCCCCCCGCCAGGAAGUCAGACUUCCUAAGCGGAACUCGCUGCA .........((((....))))....................................((((((....((((((((((.........((((((.....)))))))))))))))).)))))) (-27.46 = -27.98 + 0.52)



| Location | 8,091,360 – 8,091,480 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -45.20 |
| Consensus MFE | -36.88 |
| Energy contribution | -37.88 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8091360 120 + 27905053 CUCCCUCCUUUAUGAAGUGCAGUUUUUAGUUCCGUUUCCCUCCGCCAGGAAGUCAGACUUCCUAAGCGGAACUCGCUGCAAUAUCCUUGCAUGGGAAUGCCAGGAUUCGCGGGGUGGCAA ..((((((.........((((((....((((((((((.........((((((.....)))))))))))))))).))))))..(((((.((((....)))).)))))....)))).))... ( -43.70) >DroSec_CAF1 43140 119 + 1 CUCCAUCCUUCGUGAAGUGCAGUUUUUAGUUCUGUUUCCCCCCGCCAGGAAGUCAGACUUCCAAAGCGGAACUCGCUGCAUUAUCCUUGCAUGGGAAU-CCAGGACUCGCGGGGUGGCAA ..(((((((.(((((((......)))))((((((.(((((.((((..(((((.....)))))...)))).......((((.......)))).))))).-.)))))).)).)))))))... ( -42.00) >DroSim_CAF1 15145 120 + 1 CUCCAUCCUUCGUGAAGUGCAGUUUUUAGUUCCUUUUCUCCCCGCCAGGAAGUCAGACUUCCAAAGCGGAACUCGCUGCAUUAUCCUUGCAUGGGAAUGCCAGGACUCGCGGGGUGGCAA ..((((((..(((((((((((((....((((((..........((..(((((.....)))))...)))))))).)))))))).((((.((((....)))).)))).)))))))))))... ( -45.30) >DroEre_CAF1 18182 120 + 1 CGCCCUCCUUCAUGGACUGCAGUUUGUAGUUCAGUUUCCCCCCGCCAGGAAGUCGGACUUCCUAAGCGGAACUCGCUGCAUUAUCCUUGCAUGGGAAUGCCAGGACUCGCGGGGUGGCAA .(((........((((((((.....))))))))......((((((.((((((.....))))))..((((......))))....((((.((((....)))).))))...)))))).))).. ( -48.80) >DroYak_CAF1 11534 119 + 1 CGUCCUCCUUCAUGAAGUGCAGUUUGUAGUUCCGCUUCC-CUCGCCAGGAAGUCAGACUUCCUAAGCGGAACUCGCUGCAUUAUCCUUGCAUGGGAAUGCCAGGACUCGCGGGGUAGCAA .((..((((.(....((((((((..(.((((((((((..-......((((((.....))))))))))))))))))))))))).((((.((((....)))).))))...).))))..)).. ( -46.20) >consensus CUCCCUCCUUCAUGAAGUGCAGUUUUUAGUUCCGUUUCCCCCCGCCAGGAAGUCAGACUUCCUAAGCGGAACUCGCUGCAUUAUCCUUGCAUGGGAAUGCCAGGACUCGCGGGGUGGCAA ..((.((((.(....((((((((....((((((((((.........((((((.....)))))))))))))))).)))))))).((((.((((....)))).))))...).)))).))... (-36.88 = -37.88 + 1.00)



| Location | 8,091,360 – 8,091,480 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -41.76 |
| Consensus MFE | -33.24 |
| Energy contribution | -33.88 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8091360 120 - 27905053 UUGCCACCCCGCGAAUCCUGGCAUUCCCAUGCAAGGAUAUUGCAGCGAGUUCCGCUUAGGAAGUCUGACUUCCUGGCGGAGGGAAACGGAACUAAAAACUGCACUUCAUAAAGGAGGGAG .......(((....(((((.((((....)))).)))))..(((((..(((((((((.((((((.....)))))))))....(....))))))).....)))))............))).. ( -42.90) >DroSec_CAF1 43140 119 - 1 UUGCCACCCCGCGAGUCCUGG-AUUCCCAUGCAAGGAUAAUGCAGCGAGUUCCGCUUUGGAAGUCUGACUUCCUGGCGGGGGGAAACAGAACUAAAAACUGCACUUCACGAAGGAUGGAG .(((.........(((.(((.-.(((((.((((.......)))).......(((((..(((((.....))))).))))))))))..))).))).......)))((((((....).))))) ( -38.49) >DroSim_CAF1 15145 120 - 1 UUGCCACCCCGCGAGUCCUGGCAUUCCCAUGCAAGGAUAAUGCAGCGAGUUCCGCUUUGGAAGUCUGACUUCCUGGCGGGGAGAAAAGGAACUAAAAACUGCACUUCACGAAGGAUGGAG ...(((.((..((.(((((.((((....)))).)))))..(((((..(((((((((..(((((.....))))).)))..........)))))).....))))).....))..)).))).. ( -38.50) >DroEre_CAF1 18182 120 - 1 UUGCCACCCCGCGAGUCCUGGCAUUCCCAUGCAAGGAUAAUGCAGCGAGUUCCGCUUAGGAAGUCCGACUUCCUGGCGGGGGGAAACUGAACUACAAACUGCAGUCCAUGAAGGAGGGCG ..(((..((..((.(((((.((((....)))).)))))..(((((..(((((((((.((((((.....))))))))))..((....))))))).....))))).....))..))..))). ( -46.20) >DroYak_CAF1 11534 119 - 1 UUGCUACCCCGCGAGUCCUGGCAUUCCCAUGCAAGGAUAAUGCAGCGAGUUCCGCUUAGGAAGUCUGACUUCCUGGCGAG-GGAAGCGGAACUACAAACUGCACUUCAUGAAGGAGGACG ((((......))))(((((.((((....)))).)))))..(((((..((((((((((((((((.....))))))).....-...))))))))).....)))))((((......))))... ( -42.70) >consensus UUGCCACCCCGCGAGUCCUGGCAUUCCCAUGCAAGGAUAAUGCAGCGAGUUCCGCUUAGGAAGUCUGACUUCCUGGCGGGGGGAAACGGAACUAAAAACUGCACUUCAUGAAGGAGGGAG ...((..((..((.(((((.((((....)))).)))))..(((((..(((((((((.((((((.....))))))))))...(....).))))).....))))).....))..))..)).. (-33.24 = -33.88 + 0.64)


| Location | 8,091,400 – 8,091,520 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -43.42 |
| Consensus MFE | -41.20 |
| Energy contribution | -41.28 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8091400 120 - 27905053 CCACAAAGACCGCUGAUUACAAAGUUUGGUGCUGCUGCAGUUGCCACCCCGCGAAUCCUGGCAUUCCCAUGCAAGGAUAUUGCAGCGAGUUCCGCUUAGGAAGUCUGACUUCCUGGCGGA .........(((((.......((((..((.((((((((((((((......))))(((((.((((....)))).))))).))))))).))).))))))((((((.....))))))))))). ( -44.90) >DroSec_CAF1 43180 119 - 1 CCACAAAGACCGCUGAUUACAAAGUUUGGUGCUGCUGCAGUUGCCACCCCGCGAGUCCUGG-AUUCCCAUGCAAGGAUAAUGCAGCGAGUUCCGCUUUGGAAGUCUGACUUCCUGGCGGG .........(((((......(((((..((.(((((((((.((((......))))((((((.-((....)).).)))))..)))))).))).)))))))(((((.....))))).))))). ( -39.50) >DroSim_CAF1 15185 120 - 1 CCACAAAGACCGCUGAUUACAAGGUUUGGUGCUGCUGCAGUUGCCACCCCGCGAGUCCUGGCAUUCCCAUGCAAGGAUAAUGCAGCGAGUUCCGCUUUGGAAGUCUGACUUCCUGGCGGG .(((..(((((..((....)).))))).))).(((((((.((((......))))(((((.((((....)))).)))))..)))))))....(((((..(((((.....))))).))))). ( -44.40) >DroEre_CAF1 18222 120 - 1 CCACAAGGACCGCUGAUUACAAAGUUCGGUGCUGCUGCAGUUGCCACCCCGCGAGUCCUGGCAUUCCCAUGCAAGGAUAAUGCAGCGAGUUCCGCUUAGGAAGUCCGACUUCCUGGCGGG .....((.((((((........))..)))).))((((((.((((......))))(((((.((((....)))).)))))..)))))).....(((((.((((((.....))))))))))). ( -46.10) >DroYak_CAF1 11573 120 - 1 CCACAAAGACCGCUGAUUACAAAGUUCGGUGCUGCUGCAGUUGCUACCCCGCGAGUCCUGGCAUUCCCAUGCAAGGAUAAUGCAGCGAGUUCCGCUUAGGAAGUCUGACUUCCUGGCGAG ..........((((.......(((..(((.(((((((((.((((......))))(((((.((((....)))).)))))..)))))).))).))))))((((((.....)))))))))).. ( -42.20) >consensus CCACAAAGACCGCUGAUUACAAAGUUUGGUGCUGCUGCAGUUGCCACCCCGCGAGUCCUGGCAUUCCCAUGCAAGGAUAAUGCAGCGAGUUCCGCUUAGGAAGUCUGACUUCCUGGCGGG .........(((((.......((((..((.(((((((((.((((......))))(((((.((((....)))).)))))..)))))).))).)))))).(((((.....))))).))))). (-41.20 = -41.28 + 0.08)



| Location | 8,091,520 – 8,091,633 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -33.42 |
| Consensus MFE | -29.42 |
| Energy contribution | -29.98 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8091520 113 + 27905053 UUUAUGCUGGCAGCUCCUAAUUGAUGCAGUUAGUUGCGUGC-------UGCAACUGGAACCCUUUAAUUGAUGCUUCGGUUUGGGAAUAGAUUGCUGCUGCACUGAUAGCAACUUUGUGU ....(((((.((((..(((((((...)))))))..))((((-------.(((((((...(((...((((((....)))))).)))..))).))))....)))))).)))))......... ( -33.90) >DroSec_CAF1 43299 113 + 1 UUUAUGCUGCCAGCUCCUAAUUGAUGCAGUUAGUUGCGUGC-------UGCAACUGGAACCCUUUAAUUGAUGCUUCGGUUUGGGAAUAGAUUGCUGCUGCACUGAUAGCAACUUUGUGU ....(((((.((((..(((((((...)))))))..))((((-------.(((((((...(((...((((((....)))))).)))..))).))))....)))))).)))))......... ( -33.90) >DroSim_CAF1 15305 113 + 1 UUUAUGCUGGCAGCUCCUAAUUGAUGCAGUUAGUUGCGUGC-------UGCAACUGGAACCCUUUAAUUGAUGCUUCGGUUUGGGAAUAGAUUGCUGCUGCACUGAUAGCAACUUUGUGU ....(((((.((((..(((((((...)))))))..))((((-------.(((((((...(((...((((((....)))))).)))..))).))))....)))))).)))))......... ( -33.90) >DroEre_CAF1 18342 119 + 1 UUUAUGCUGGCAGCUCCUAAUUGAUGCAGUUAGCUGCGUGCAGUGAGUUGCGACUGGAACCCUUUAAUUGAUGCUUCGG-UUGGGAAUAGAUUGCUGCAGCACUGAUAGCAACUUUGUUU .........(((((((...((((((((((....)))))).)))))))))))........(((...((((((....))))-)))))((((((((((((((....)).)))))).)))))). ( -35.80) >DroYak_CAF1 11693 118 + 1 UUUAUGCUGGCAGCUCCUAAUUGAUGCAGUUAGUUGCGUGCAGUUAGUUGCGACUGGAACCCUUUAAUUGAUGCUUCGC-UUGGGAAUAGAUUGGU-UGGCACUGAUAGCAACUUUGUGU ....(((..((.((..(((((((((((((....)))))).)))))))..))..(((...(((......(((....))).-..)))..)))....))-..))).................. ( -29.60) >consensus UUUAUGCUGGCAGCUCCUAAUUGAUGCAGUUAGUUGCGUGC_______UGCAACUGGAACCCUUUAAUUGAUGCUUCGGUUUGGGAAUAGAUUGCUGCUGCACUGAUAGCAACUUUGUGU ....(((((.((((..(((((((...)))))))..))((((........(((((((...(((...((((((....)))))).)))..))).))))....)))))).)))))......... (-29.42 = -29.98 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:33 2006