
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,090,787 – 8,090,964 |
| Length | 177 |
| Max. P | 0.999706 |

| Location | 8,090,787 – 8,090,884 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.67 |
| Mean single sequence MFE | -31.56 |
| Consensus MFE | -22.42 |
| Energy contribution | -24.78 |
| Covariance contribution | 2.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.71 |
| SVM decision value | 3.30 |
| SVM RNA-class probability | 0.998950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8090787 97 + 27905053 UGGCUUUUGUGCUAUUUGGAACGAGUUGUCAAAUAAAAGCGGCAGCCGGCGAAAUAGAAGAAGGAGCCAGGGA-----------------------AGCCUGCCGAAGGGGGCAGGAGGA ((((((((...((((((.(..((.((((((..........)))))))).).))))))....))))))))....-----------------------..((((((......)))))).... ( -35.80) >DroSec_CAF1 42576 83 + 1 UGGCUUUUGUGCUAUUUGGAACGAGUUGUCAAAUAAAAGCGGCAGCCGGCGGAAUAGGAGAAGGAGCCAGGGA-----------------------AG--------------GAGGAGGA ((((((((.(.((((((.(..((.((((((..........)))))))).).)))))).)..))))))))....-----------------------..--------------........ ( -24.40) >DroSim_CAF1 14581 83 + 1 UGGCUUUUGUGCUAUUUGGAACGAGUUGUCAAAUAAAAGCGGCAGCCGGCGGAAUAGGAGAAGGAGCCAGGGA-----------------------AG--------------GAGGAGGA ((((((((.(.((((((.(..((.((((((..........)))))))).).)))))).)..))))))))....-----------------------..--------------........ ( -24.40) >DroEre_CAF1 17551 120 + 1 UGGCUUUUGUGCUAUUUGGAACGAGUUGUCAAAUAAAAGCGGCAGCCGGCGGAGAAGUAGAAGGAUCCAGGAACCAGUAACCAGUAACCAGGAGCCAUCCUGCCGAAGGCGGCAGGAGGA (((((((((((((..((((..((.((((((..........))))))))..(((............))).....)))).....))))..)))))))))((((((((....))))))))... ( -41.70) >DroYak_CAF1 10946 96 + 1 UGGCUUUUGUGCUAUUUGGAACGAGUUGUCAAAUAAAAGCGGCAGCCGGCGGA-AAGUAGAAGGAACCGGGGA-----------------------GUCCUGCCGAAGGCGGCAGGAGGA .((.((((...((((((....((.((((((..........)))))))).....-))))))..)))))).....-----------------------.((((((((....))))))))... ( -31.50) >consensus UGGCUUUUGUGCUAUUUGGAACGAGUUGUCAAAUAAAAGCGGCAGCCGGCGGAAUAGGAGAAGGAGCCAGGGA_______________________AGCCUGCCGAAGG_GGCAGGAGGA ((((((((...((((((.(..((.((((((..........)))))))).).))))))....)))))))).............................((((((......)))))).... (-22.42 = -24.78 + 2.36)



| Location | 8,090,787 – 8,090,884 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.67 |
| Mean single sequence MFE | -21.34 |
| Consensus MFE | -13.22 |
| Energy contribution | -15.10 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8090787 97 - 27905053 UCCUCCUGCCCCCUUCGGCAGGCU-----------------------UCCCUGGCUCCUUCUUCUAUUUCGCCGGCUGCCGCUUUUAUUUGACAACUCGUUCCAAAUAGCACAAAAGCCA ....((((((......))))))..-----------------------.....(((...............)))((((...(((...(((((...........)))))))).....)))). ( -22.16) >DroSec_CAF1 42576 83 - 1 UCCUCCUC--------------CU-----------------------UCCCUGGCUCCUUCUCCUAUUCCGCCGGCUGCCGCUUUUAUUUGACAACUCGUUCCAAAUAGCACAAAAGCCA ........--------------..-----------------------.....(((...............)))((((...(((...(((((...........)))))))).....)))). ( -11.96) >DroSim_CAF1 14581 83 - 1 UCCUCCUC--------------CU-----------------------UCCCUGGCUCCUUCUCCUAUUCCGCCGGCUGCCGCUUUUAUUUGACAACUCGUUCCAAAUAGCACAAAAGCCA ........--------------..-----------------------.....(((...............)))((((...(((...(((((...........)))))))).....)))). ( -11.96) >DroEre_CAF1 17551 120 - 1 UCCUCCUGCCGCCUUCGGCAGGAUGGCUCCUGGUUACUGGUUACUGGUUCCUGGAUCCUUCUACUUCUCCGCCGGCUGCCGCUUUUAUUUGACAACUCGUUCCAAAUAGCACAAAAGCCA ...((((((((....))))))))(((((...(((..(((((....((....((((....)))).....)))))))..)))(((...(((((...........)))))))).....))))) ( -36.40) >DroYak_CAF1 10946 96 - 1 UCCUCCUGCCGCCUUCGGCAGGAC-----------------------UCCCCGGUUCCUUCUACUU-UCCGCCGGCUGCCGCUUUUAUUUGACAACUCGUUCCAAAUAGCACAAAAGCCA ...((((((((....)))))))).-----------------------....(((............-.)))..((((...(((...(((((...........)))))))).....)))). ( -24.22) >consensus UCCUCCUGCC_CCUUCGGCAGGCU_______________________UCCCUGGCUCCUUCUACUAUUCCGCCGGCUGCCGCUUUUAUUUGACAACUCGUUCCAAAUAGCACAAAAGCCA ....((((((......))))))..............................(((...............)))((((...(((...(((((...........)))))))).....)))). (-13.22 = -15.10 + 1.88)



| Location | 8,090,827 – 8,090,924 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.29 |
| Mean single sequence MFE | -43.16 |
| Consensus MFE | -31.15 |
| Energy contribution | -33.03 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -4.44 |
| Structure conservation index | 0.72 |
| SVM decision value | 3.92 |
| SVM RNA-class probability | 0.999706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8090827 97 + 27905053 GGCAGCCGGCGAAAUAGAAGAAGGAGCCAGGGA-----------------------AGCCUGCCGAAGGGGGCAGGAGGAGCUGGAUCGUUGACUUGGCUGGCUGGCUGCCAUCAAAAUG ((((((((((.(..(((...((.((.((((...-----------------------..((((((......)))))).....)))).)).))...)))..).))))))))))......... ( -43.70) >DroSec_CAF1 42616 83 + 1 GGCAGCCGGCGGAAUAGGAGAAGGAGCCAGGGA-----------------------AG--------------GAGGAGGAGCUGGAUCGUUGACUUGGCUGGCUGGCUGCCAUCAAAAUG ((((((((((.(..((((..((.((.((((...-----------------------..--------------.........)))).)).))..))))..).))))))))))......... ( -33.24) >DroSim_CAF1 14621 83 + 1 GGCAGCCGGCGGAAUAGGAGAAGGAGCCAGGGA-----------------------AG--------------GAGGAGGAGCUGGAUCGUUGACUUGGCUGGCUGGCUGCCAUCAAAAUG ((((((((((.(..((((..((.((.((((...-----------------------..--------------.........)))).)).))..))))..).))))))))))......... ( -33.24) >DroEre_CAF1 17591 120 + 1 GGCAGCCGGCGGAGAAGUAGAAGGAUCCAGGAACCAGUAACCAGUAACCAGGAGCCAUCCUGCCGAAGGCGGCAGGAGGAGCAGGAUCGUUGACUUGGCUGGCUGGCUGCCAUCAAAAUG ((((((((((((..((((..((.(((((.((.........)).........(..((.((((((((....))))))))))..).))))).)).))))..)).))))))))))......... ( -56.30) >DroYak_CAF1 10986 96 + 1 GGCAGCCGGCGGA-AAGUAGAAGGAACCGGGGA-----------------------GUCCUGCCGAAGGCGGCAGGAGGAGCUGGAUCGUUGACUUGGCUGGCUGGCUGCCAUCAAAAUG ((((((((((((.-((((..((.((.((((...-----------------------.((((((((....))))))))....)))).)).)).))))..)).))))))))))......... ( -49.30) >consensus GGCAGCCGGCGGAAUAGGAGAAGGAGCCAGGGA_______________________AGCCUGCCGAAGG_GGCAGGAGGAGCUGGAUCGUUGACUUGGCUGGCUGGCUGCCAUCAAAAUG ((((((((((.(.............((((((...........................((((((......))))))..((......)).....))))))).))))))))))......... (-31.15 = -33.03 + 1.88)

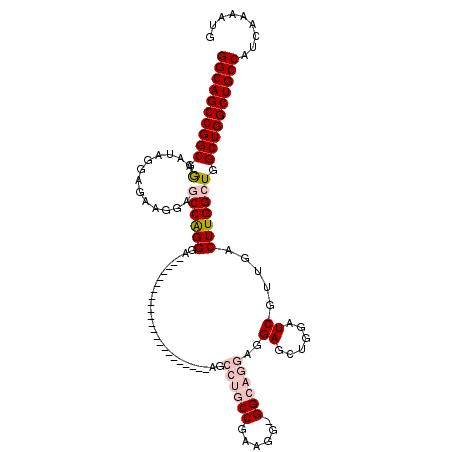
| Location | 8,090,827 – 8,090,924 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.29 |
| Mean single sequence MFE | -30.16 |
| Consensus MFE | -20.58 |
| Energy contribution | -22.38 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8090827 97 - 27905053 CAUUUUGAUGGCAGCCAGCCAGCCAAGUCAACGAUCCAGCUCCUCCUGCCCCCUUCGGCAGGCU-----------------------UCCCUGGCUCCUUCUUCUAUUUCGCCGGCUGCC .........(((((((.((.............((.((((.....((((((......))))))..-----------------------...)))).)).............)).))))))) ( -33.07) >DroSec_CAF1 42616 83 - 1 CAUUUUGAUGGCAGCCAGCCAGCCAAGUCAACGAUCCAGCUCCUCCUC--------------CU-----------------------UCCCUGGCUCCUUCUCCUAUUCCGCCGGCUGCC .........(((((((((((((..(((.....((......))......--------------))-----------------------)..)))))).................))))))) ( -19.92) >DroSim_CAF1 14621 83 - 1 CAUUUUGAUGGCAGCCAGCCAGCCAAGUCAACGAUCCAGCUCCUCCUC--------------CU-----------------------UCCCUGGCUCCUUCUCCUAUUCCGCCGGCUGCC .........(((((((((((((..(((.....((......))......--------------))-----------------------)..)))))).................))))))) ( -19.92) >DroEre_CAF1 17591 120 - 1 CAUUUUGAUGGCAGCCAGCCAGCCAAGUCAACGAUCCUGCUCCUCCUGCCGCCUUCGGCAGGAUGGCUCCUGGUUACUGGUUACUGGUUCCUGGAUCCUUCUACUUCUCCGCCGGCUGCC .........(((((((.((.....((((....(((((.(((..((((((((....)))))))).)))....((..((..(...)..)).)).))))).....))))....)).))))))) ( -44.50) >DroYak_CAF1 10986 96 - 1 CAUUUUGAUGGCAGCCAGCCAGCCAAGUCAACGAUCCAGCUCCUCCUGCCGCCUUCGGCAGGAC-----------------------UCCCCGGUUCCUUCUACUU-UCCGCCGGCUGCC .........(((((((.((.....((((....((.((.(....((((((((....)))))))).-----------------------...).)).)).....))))-...)).))))))) ( -33.40) >consensus CAUUUUGAUGGCAGCCAGCCAGCCAAGUCAACGAUCCAGCUCCUCCUGCC_CCUUCGGCAGGCU_______________________UCCCUGGCUCCUUCUACUAUUCCGCCGGCUGCC .........(((((((.((.(((...(((...)))...)))...((((((......))))))................................................)).))))))) (-20.58 = -22.38 + 1.80)



| Location | 8,090,860 – 8,090,964 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.04 |
| Mean single sequence MFE | -42.20 |
| Consensus MFE | -35.53 |
| Energy contribution | -37.33 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8090860 104 + 27905053 ----------------AGCCUGCCGAAGGGGGCAGGAGGAGCUGGAUCGUUGACUUGGCUGGCUGGCUGCCAUCAAAAUGGCGUCUGGCCCGGCAAGGACUUUAAAAAGCUGCUCGGAAA ----------------...(((((......)))))(((.((((..........((((.(.(((..(.((((((....)))))).)..))).).))))..........)))).)))..... ( -44.25) >DroSec_CAF1 42649 90 + 1 ----------------AG--------------GAGGAGGAGCUGGAUCGUUGACUUGGCUGGCUGGCUGCCAUCAAAAUGGCGUCUGGCCCAGCAAGGACUUUAAAAAGCUGCUCGGAAA ----------------..--------------...(((.((((..........((((.(((((..(.((((((....)))))).)..).))))))))..........)))).)))..... ( -33.85) >DroSim_CAF1 14654 90 + 1 ----------------AG--------------GAGGAGGAGCUGGAUCGUUGACUUGGCUGGCUGGCUGCCAUCAAAAUGGCGUCUGGCCCGGCAAGGACUUUAAAAAGCUGCUCGGAAA ----------------..--------------...(((.((((..........((((.(.(((..(.((((((....)))))).)..))).).))))..........)))).)))..... ( -35.55) >DroEre_CAF1 17631 120 + 1 CCAGUAACCAGGAGCCAUCCUGCCGAAGGCGGCAGGAGGAGCAGGAUCGUUGACUUGGCUGGCUGGCUGCCAUCAAAAUGGCGUCUGGCCCGGCAAGGACUUUAAAAAGCUGCUCGGAAA ((........))..((.((((((((....)))))))).((((((.........((((.(.(((..(.((((((....)))))).)..))).).))))..(((....)))))))))))... ( -51.20) >DroYak_CAF1 11018 104 + 1 ----------------GUCCUGCCGAAGGCGGCAGGAGGAGCUGGAUCGUUGACUUGGCUGGCUGGCUGCCAUCAAAAUGGCGUCUGGCCCGGCAAGGACUUUAAAAAGCUGCUCGGAAA ----------------...((((((....))))))(((.((((..........((((.(.(((..(.((((((....)))))).)..))).).))))..........)))).)))..... ( -46.15) >consensus ________________AGCCUGCCGAAGG_GGCAGGAGGAGCUGGAUCGUUGACUUGGCUGGCUGGCUGCCAUCAAAAUGGCGUCUGGCCCGGCAAGGACUUUAAAAAGCUGCUCGGAAA ...................(((((......)))))(((.((((..........((((.(.(((..(.((((((....)))))).)..))).).))))..........)))).)))..... (-35.53 = -37.33 + 1.80)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:25 2006