
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,088,290 – 8,088,410 |
| Length | 120 |
| Max. P | 0.999578 |

| Location | 8,088,290 – 8,088,410 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -41.14 |
| Consensus MFE | -39.22 |
| Energy contribution | -40.06 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8088290 120 + 27905053 AUGAUUUUUGUAUUCAAAUGCGGAUUGCUCCAACCAGUUUGGGCAGUUCCAAUACCACCCAAGGGAGUGUCCACUGAAUGGCAAGGCGCGCAUCCUGCUGACGGCAGGAUUGGCCCAUCG ..(((..(((((((((...(.((((.(((((......((((((..((......))..))))))))))))))).)))))).))))(((.((.(((((((.....)))))))))))).))). ( -38.80) >DroSec_CAF1 13164 120 + 1 AUGAUUUUUGUAUUCAAAUGCGGAUUGCUCCAACCAGUUUGGGCAGUUCCCUUACCACCCAAGGGAGUGCCCACUGAAUGGCAAGGCGCGCAUCCUGCUGACGGCAGGAUUGGCCCAUCG ..(((.(((((((....)))))))(((((.....((((..(((((.(((((((.......))))))))))))))))...)))))(((.((.(((((((.....)))))))))))).))). ( -44.50) >DroSim_CAF1 11984 120 + 1 AUGAUUUUUGUAUUCAAAUGCGGAUUGCUCCAACCAGUUUGGGCAGUUCCCUUACCACCCAAGGGAGUGUCCACUGAAUGGCAAGGCGCGCAUCCUGCUGACGGCAGGAUUGGCCCAACG ......(((((((....)))))))(((((.....((((..(((((.(((((((.......))))))))))))))))...)))))(((.((.(((((((.....))))))))))))..... ( -41.70) >DroEre_CAF1 14998 120 + 1 AUGAUUUUUGUAUUCAAAUGCGGAUUGCUCCAACCAGUUUGGGCAGUUCCCUCAGCACCCAAGGGAGUGUCCACUGAAUGGCAAGGCGCGCAUCCUGCUGACGGCAGGAUUGGCCCAUCG ..(((.(((((((....)))))))(((((.....((((..(((((.((((((.........)))))))))))))))...)))))(((.((.(((((((.....)))))))))))).))). ( -40.30) >DroYak_CAF1 8352 120 + 1 AUGAUUUUUGUAUUCAAAUGCGGAUUGCUCCAACCAGUUUGGGCAGUUCCCUCUGCACCCAAGGGAGUGUCCCCUGAAUGGCAAGGCGCGCAUCCUGCUGACGGCAGGAUUGGCCCAUCG ..(((..((((((((...((((((((((.((((.....)))))))).....))))))....((((......)))))))).))))(((.((.(((((((.....)))))))))))).))). ( -40.40) >consensus AUGAUUUUUGUAUUCAAAUGCGGAUUGCUCCAACCAGUUUGGGCAGUUCCCUUACCACCCAAGGGAGUGUCCACUGAAUGGCAAGGCGCGCAUCCUGCUGACGGCAGGAUUGGCCCAUCG ......(((((((....)))))))(((((.....((((..(((((.(((((((.......))))))))))))))))...)))))(((.((.(((((((.....))))))))))))..... (-39.22 = -40.06 + 0.84)



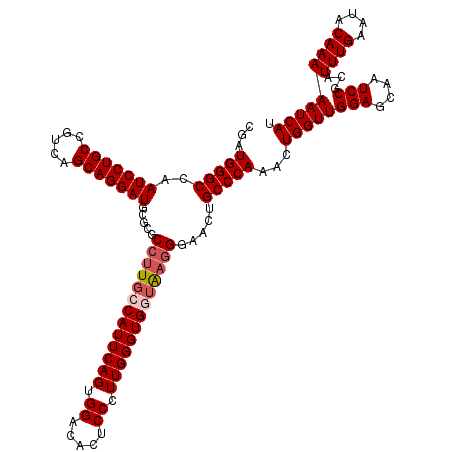
| Location | 8,088,290 – 8,088,410 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -45.18 |
| Consensus MFE | -42.26 |
| Energy contribution | -43.02 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.74 |
| SVM RNA-class probability | 0.999578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8088290 120 - 27905053 CGAUGGGCCAAUCCUGCCGUCAGCAGGAUGCGCGCCUUGCCAUUCAGUGGACACUCCCUUGGGUGGUAUUGGAACUGCCCAAACUGGUUGGAGCAAUCCGCAUUUGAAUACAAAAAUCAU ...(((((..(((((((.....)))))))..(..((.((((((((((.((.....)).))))))))))..))..).)))))...((((((((....)))...((((....))))))))). ( -42.20) >DroSec_CAF1 13164 120 - 1 CGAUGGGCCAAUCCUGCCGUCAGCAGGAUGCGCGCCUUGCCAUUCAGUGGGCACUCCCUUGGGUGGUAAGGGAACUGCCCAAACUGGUUGGAGCAAUCCGCAUUUGAAUACAAAAAUCAU ...(((((..(((((((.....))))))).....(((((((((((((.(((....)))))))))))))))).....)))))...((((((((....)))...((((....))))))))). ( -48.20) >DroSim_CAF1 11984 120 - 1 CGUUGGGCCAAUCCUGCCGUCAGCAGGAUGCGCGCCUUGCCAUUCAGUGGACACUCCCUUGGGUGGUAAGGGAACUGCCCAAACUGGUUGGAGCAAUCCGCAUUUGAAUACAAAAAUCAU ..((((((..(((((((.....))))))).....(((((((((((((.((.....)).))))))))))))).....))))))..((((((((....)))...((((....))))))))). ( -47.90) >DroEre_CAF1 14998 120 - 1 CGAUGGGCCAAUCCUGCCGUCAGCAGGAUGCGCGCCUUGCCAUUCAGUGGACACUCCCUUGGGUGCUGAGGGAACUGCCCAAACUGGUUGGAGCAAUCCGCAUUUGAAUACAAAAAUCAU .((((((((.(((((((.....)))))))..).))))....((((((((((...(((((..(...)..)))))..(((((((.....)))).))).)))))...)))))......))).. ( -42.20) >DroYak_CAF1 8352 120 - 1 CGAUGGGCCAAUCCUGCCGUCAGCAGGAUGCGCGCCUUGCCAUUCAGGGGACACUCCCUUGGGUGCAGAGGGAACUGCCCAAACUGGUUGGAGCAAUCCGCAUUUGAAUACAAAAAUCAU ...(((((..(((((((.....))))))).....((((..((((((((((.....))))))))))..)))).....)))))...((((((((....)))...((((....))))))))). ( -45.40) >consensus CGAUGGGCCAAUCCUGCCGUCAGCAGGAUGCGCGCCUUGCCAUUCAGUGGACACUCCCUUGGGUGGUAAGGGAACUGCCCAAACUGGUUGGAGCAAUCCGCAUUUGAAUACAAAAAUCAU ...(((((..(((((((.....))))))).....(((((((((((((.((.....)).))))))))))))).....)))))...((((((((....)))...((((....))))))))). (-42.26 = -43.02 + 0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:15 2006