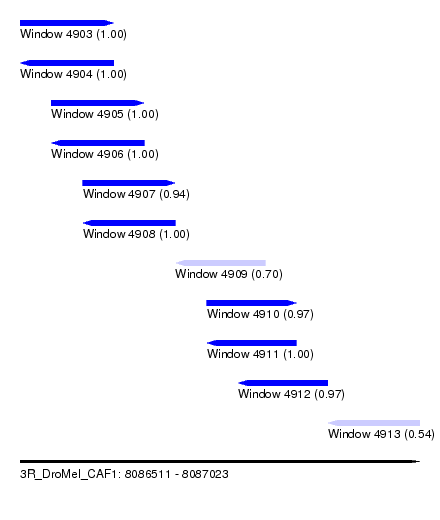
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,086,511 – 8,087,023 |
| Length | 512 |
| Max. P | 0.999969 |

| Location | 8,086,511 – 8,086,631 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.92 |
| Mean single sequence MFE | -40.76 |
| Consensus MFE | -37.36 |
| Energy contribution | -37.92 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.08 |
| SVM RNA-class probability | 0.999790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8086511 120 + 27905053 GCGGUUUUAAGGCCAUUCGCAACACCCUCGACGAGUGGUCGCUGAAAAGUUUAAGCGAUUAGCGCAAUUUAGCGAAAAUAACAUGCGAAAUUGCGCGCUUGAACGCCUUUUCAAUGGCAC ....(((((.((((((((((.........).)))))))))..))))).((((((((.....(((((((((.(((.........))).)))))))))))))))))(((........))).. ( -41.10) >DroSec_CAF1 11441 120 + 1 GCGGUUUUAAGGCCAUUCGCCACACCCUCGACGAGUGGUCGCUGAAAAGUUUAAGCGAUUAGCGCAAUUUUGCGAAAAUAACAUGCGAAAUUGCGCGCUUGAACGCCUUUUCAAUGGCAC ....(((((.(((((((((.(........).)))))))))..))))).((((((((.....(((((((((((((.........)))))))))))))))))))))(((........))).. ( -45.70) >DroSim_CAF1 10276 120 + 1 GCGGUUUUAAGGCCAUUCGCCACACCCUCGACGAGUGGUCGCUGAAAAGUUUAAGCGAUUAGCGCAAUUUAGCGAAAAUAACAUGCGAAAUUGCGCGCUUGAACGCCUCUUCAAUGGCAC ....(((((.(((((((((.(........).)))))))))..))))).((((((((.....(((((((((.(((.........))).)))))))))))))))))(((........))).. ( -41.90) >DroEre_CAF1 13199 120 + 1 GUGGUUGCAAUGCCAUUCGCCCCACCCUCGACGAGUGGUCGCUGAAAAGUUUAAGCGAUUAGCGCAAUUUAGCGAAAAUAACAUGCGAAAUUGCGCGCUUGAACGCCUUUUCAAUGGCAC ......((...((((((((.(........).)))))))).))......((((((((.....(((((((((.(((.........))).)))))))))))))))))(((........))).. ( -37.80) >DroYak_CAF1 6557 120 + 1 GUGGUUUCACCGCCAUUCGCCCCACCCUCGACGAGUGGUCGCUGAAAAGUUUAAGCGAUUAGCGCAAUUUAGCGAAAAUAACAUGCGAAAUUGCGCGAUUGAACGCCUUUUCAAUGGCAC ((((.....))))((((((.(........).))))))((((.(((((((......(((((.(((((((((.(((.........))).)))))))))))))).....))))))).)))).. ( -37.30) >consensus GCGGUUUUAAGGCCAUUCGCCACACCCUCGACGAGUGGUCGCUGAAAAGUUUAAGCGAUUAGCGCAAUUUAGCGAAAAUAACAUGCGAAAUUGCGCGCUUGAACGCCUUUUCAAUGGCAC .((((.....(((((((((.(........).)))))))))))))....((((((((.....(((((((((.(((.........))).)))))))))))))))))(((........))).. (-37.36 = -37.92 + 0.56)


| Location | 8,086,511 – 8,086,631 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.92 |
| Mean single sequence MFE | -40.72 |
| Consensus MFE | -36.12 |
| Energy contribution | -36.16 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8086511 120 - 27905053 GUGCCAUUGAAAAGGCGUUCAAGCGCGCAAUUUCGCAUGUUAUUUUCGCUAAAUUGCGCUAAUCGCUUAAACUUUUCAGCGACCACUCGUCGAGGGUGUUGCGAAUGGCCUUAAAACCGC ..((((((((((((((......))(((((((((.((...........)).)))))))))............)))))).(((((.((((.....))))))))).))))))........... ( -40.30) >DroSec_CAF1 11441 120 - 1 GUGCCAUUGAAAAGGCGUUCAAGCGCGCAAUUUCGCAUGUUAUUUUCGCAAAAUUGCGCUAAUCGCUUAAACUUUUCAGCGACCACUCGUCGAGGGUGUGGCGAAUGGCCUUAAAACCGC ..((((((((((((((......))(((((((((.((...........)).)))))))))............)))))).((.((.((((.....)))))).)).))))))........... ( -38.60) >DroSim_CAF1 10276 120 - 1 GUGCCAUUGAAGAGGCGUUCAAGCGCGCAAUUUCGCAUGUUAUUUUCGCUAAAUUGCGCUAAUCGCUUAAACUUUUCAGCGACCACUCGUCGAGGGUGUGGCGAAUGGCCUUAAAACCGC ..((((((((((((((......))(((((((((.((...........)).)))))))))............)))))).((.((.((((.....)))))).)).))))))........... ( -38.70) >DroEre_CAF1 13199 120 - 1 GUGCCAUUGAAAAGGCGUUCAAGCGCGCAAUUUCGCAUGUUAUUUUCGCUAAAUUGCGCUAAUCGCUUAAACUUUUCAGCGACCACUCGUCGAGGGUGGGGCGAAUGGCAUUGCAACCAC ((((((((((((((((......))(((((((((.((...........)).)))))))))............)))))).((..((((((.....)))))).)).))))))))......... ( -44.00) >DroYak_CAF1 6557 120 - 1 GUGCCAUUGAAAAGGCGUUCAAUCGCGCAAUUUCGCAUGUUAUUUUCGCUAAAUUGCGCUAAUCGCUUAAACUUUUCAGCGACCACUCGUCGAGGGUGGGGCGAAUGGCGGUGAAACCAC ..(((((((((((((((.......(((((((((.((...........)).)))))))))....))).....)))))).((..((((((.....)))))).)).))))))((.....)).. ( -42.00) >consensus GUGCCAUUGAAAAGGCGUUCAAGCGCGCAAUUUCGCAUGUUAUUUUCGCUAAAUUGCGCUAAUCGCUUAAACUUUUCAGCGACCACUCGUCGAGGGUGUGGCGAAUGGCCUUAAAACCGC ..((((((((((((((......))(((((((((.((...........)).)))))))))............)))))).((...(((((.....)))))..)).))))))........... (-36.12 = -36.16 + 0.04)



| Location | 8,086,551 – 8,086,670 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.82 |
| Mean single sequence MFE | -36.06 |
| Consensus MFE | -35.22 |
| Energy contribution | -35.18 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.80 |
| SVM RNA-class probability | 0.999951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8086551 119 + 27905053 GCUGAAAAGUUUAAGCGAUUAGCGCAAUUUAGCGAAAAUAACAUGCGAAAUUGCGCGCUUGAACGCCUUUUCAAUGGCACUCAGAUACAAAAGCAGCCACAGUUGCAGAUAGUGC-AAAA .((((...((((((((.....(((((((((.(((.........))).)))))))))))))))))(((........)))..))))........(((((....))))).........-.... ( -36.60) >DroSec_CAF1 11481 119 + 1 GCUGAAAAGUUUAAGCGAUUAGCGCAAUUUUGCGAAAAUAACAUGCGAAAUUGCGCGCUUGAACGCCUUUUCAAUGGCACUCAGAUACAAAAGCAGCCACAGUUGCAGAUAGUGC-AAAA .((((...((((((((.....(((((((((((((.........)))))))))))))))))))))(((........)))..))))........(((((....))))).........-.... ( -40.40) >DroSim_CAF1 10316 119 + 1 GCUGAAAAGUUUAAGCGAUUAGCGCAAUUUAGCGAAAAUAACAUGCGAAAUUGCGCGCUUGAACGCCUCUUCAAUGGCACUCAGAUACAAAAGCAGCCACAGUUGCAGAUAGUGC-ACAA .((((...((((((((.....(((((((((.(((.........))).)))))))))))))))))(((........)))..))))........(((((....))))).........-.... ( -36.60) >DroEre_CAF1 13239 120 + 1 GCUGAAAAGUUUAAGCGAUUAGCGCAAUUUAGCGAAAAUAACAUGCGAAAUUGCGCGCUUGAACGCCUUUUCAAUGGCACUCAGAUACAAAAGCGGCCACAGUUGCAGAUAGUGCGAAAA .((((...((((((((.....(((((((((.(((.........))).)))))))))))))))))(((........)))..))))........(((((....))))).............. ( -35.80) >DroYak_CAF1 6597 119 + 1 GCUGAAAAGUUUAAGCGAUUAGCGCAAUUUAGCGAAAAUAACAUGCGAAAUUGCGCGAUUGAACGCCUUUUCAAUGGCACUCAGAUACAAAAGCGGCCACAGUUGCAGAUAGUGC-AAAA .((((...((((((.......(((((((((.(((.........))).)))))))))..))))))(((........)))..))))........(((((....))))).........-.... ( -30.90) >consensus GCUGAAAAGUUUAAGCGAUUAGCGCAAUUUAGCGAAAAUAACAUGCGAAAUUGCGCGCUUGAACGCCUUUUCAAUGGCACUCAGAUACAAAAGCAGCCACAGUUGCAGAUAGUGC_AAAA .((((...((((((((.....(((((((((.(((.........))).)))))))))))))))))(((........)))..))))........(((((....))))).............. (-35.22 = -35.18 + -0.04)



| Location | 8,086,551 – 8,086,670 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.82 |
| Mean single sequence MFE | -34.34 |
| Consensus MFE | -33.02 |
| Energy contribution | -33.06 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.06 |
| SVM RNA-class probability | 0.999781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8086551 119 - 27905053 UUUU-GCACUAUCUGCAACUGUGGCUGCUUUUGUAUCUGAGUGCCAUUGAAAAGGCGUUCAAGCGCGCAAUUUCGCAUGUUAUUUUCGCUAAAUUGCGCUAAUCGCUUAAACUUUUCAGC ..((-(((.....)))))..(((((.((((........)))))))))(((((((((......))(((((((((.((...........)).)))))))))............))))))).. ( -34.70) >DroSec_CAF1 11481 119 - 1 UUUU-GCACUAUCUGCAACUGUGGCUGCUUUUGUAUCUGAGUGCCAUUGAAAAGGCGUUCAAGCGCGCAAUUUCGCAUGUUAUUUUCGCAAAAUUGCGCUAAUCGCUUAAACUUUUCAGC ..((-(((.....)))))..(((((.((((........)))))))))(((((((((......))(((((((((.((...........)).)))))))))............))))))).. ( -34.70) >DroSim_CAF1 10316 119 - 1 UUGU-GCACUAUCUGCAACUGUGGCUGCUUUUGUAUCUGAGUGCCAUUGAAGAGGCGUUCAAGCGCGCAAUUUCGCAUGUUAUUUUCGCUAAAUUGCGCUAAUCGCUUAAACUUUUCAGC .((.-(((((...(((((..((....))..)))))....)))))))((((((((((......))(((((((((.((...........)).)))))))))............)))))))). ( -35.10) >DroEre_CAF1 13239 120 - 1 UUUUCGCACUAUCUGCAACUGUGGCCGCUUUUGUAUCUGAGUGCCAUUGAAAAGGCGUUCAAGCGCGCAAUUUCGCAUGUUAUUUUCGCUAAAUUGCGCUAAUCGCUUAAACUUUUCAGC .....(((((...(((((..((....))..)))))....)))))..((((((((((......))(((((((((.((...........)).)))))))))............)))))))). ( -34.70) >DroYak_CAF1 6597 119 - 1 UUUU-GCACUAUCUGCAACUGUGGCCGCUUUUGUAUCUGAGUGCCAUUGAAAAGGCGUUCAAUCGCGCAAUUUCGCAUGUUAUUUUCGCUAAAUUGCGCUAAUCGCUUAAACUUUUCAGC ..((-(((.....)))))..(((((.((((........)))))))))((((((((((.......(((((((((.((...........)).)))))))))....))).....))))))).. ( -32.50) >consensus UUUU_GCACUAUCUGCAACUGUGGCUGCUUUUGUAUCUGAGUGCCAUUGAAAAGGCGUUCAAGCGCGCAAUUUCGCAUGUUAUUUUCGCUAAAUUGCGCUAAUCGCUUAAACUUUUCAGC .....(((((...(((((..((....))..)))))....)))))..((((((((((......))(((((((((.((...........)).)))))))))............)))))))). (-33.02 = -33.06 + 0.04)



| Location | 8,086,591 – 8,086,710 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.65 |
| Mean single sequence MFE | -36.94 |
| Consensus MFE | -33.92 |
| Energy contribution | -33.88 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.940304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8086591 119 + 27905053 ACAUGCGAAAUUGCGCGCUUGAACGCCUUUUCAAUGGCACUCAGAUACAAAAGCAGCCACAGUUGCAGAUAGUGC-AAAACAGGGAAUCCCGCUGAGCUGAGCCGCAUUUUGAGCGCACU ....(((....)))(((((..((.(((........))).(((((.........((((.....(((((.....)))-))....(((...)))))))..)))))......))..)))))... ( -35.70) >DroSec_CAF1 11521 119 + 1 ACAUGCGAAAUUGCGCGCUUGAACGCCUUUUCAAUGGCACUCAGAUACAAAAGCAGCCACAGUUGCAGAUAGUGC-AAAACAGGGAAUCCCGCUGAGCUGAGCCGCAUUUUGAGCGCACU ....(((....)))(((((..((.(((........))).(((((.........((((.....(((((.....)))-))....(((...)))))))..)))))......))..)))))... ( -35.70) >DroSim_CAF1 10356 119 + 1 ACAUGCGAAAUUGCGCGCUUGAACGCCUCUUCAAUGGCACUCAGAUACAAAAGCAGCCACAGUUGCAGAUAGUGC-ACAACAGGGAAUCCCGCUGAGCUGAGCCGCAUUUUGAGCGCACU ....(((....)))(((((..((.(((........))).(((((........(((((....))))).........-....(((((....)).)))..)))))......))..)))))... ( -35.70) >DroEre_CAF1 13279 120 + 1 ACAUGCGAAAUUGCGCGCUUGAACGCCUUUUCAAUGGCACUCAGAUACAAAAGCGGCCACAGUUGCAGAUAGUGCGAAAAAAGGGAAUCCCGCUGAGCUGAGCCGCAUUUUGAGCGCACU ....(((....)))(((((..((.(((........)))..............(((((..((((((((.....))).......(((...)))....))))).)))))..))..)))))... ( -39.20) >DroYak_CAF1 6637 119 + 1 ACAUGCGAAAUUGCGCGAUUGAACGCCUUUUCAAUGGCACUCAGAUACAAAAGCGGCCACAGUUGCAGAUAGUGC-AAAAAAGGGAAUCCCGCUGAGCUGAGCCGCAUUUUGAGCGCACU ...........(((((..((((..(((........)))..))))...((((((((((..((((((((.....)))-......(((...)))....))))).))))).))))).))))).. ( -38.40) >consensus ACAUGCGAAAUUGCGCGCUUGAACGCCUUUUCAAUGGCACUCAGAUACAAAAGCAGCCACAGUUGCAGAUAGUGC_AAAACAGGGAAUCCCGCUGAGCUGAGCCGCAUUUUGAGCGCACU ...((((.((.((((.((((....(((........))).(((((........(((((....)))))................(((...))).)))))..))))))))..))...)))).. (-33.92 = -33.88 + -0.04)



| Location | 8,086,591 – 8,086,710 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.65 |
| Mean single sequence MFE | -43.04 |
| Consensus MFE | -40.54 |
| Energy contribution | -40.90 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.62 |
| SVM RNA-class probability | 0.999461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8086591 119 - 27905053 AGUGCGCUCAAAAUGCGGCUCAGCUCAGCGGGAUUCCCUGUUUU-GCACUAUCUGCAACUGUGGCUGCUUUUGUAUCUGAGUGCCAUUGAAAAGGCGUUCAAGCGCGCAAUUUCGCAUGU .((((((((((((.(((((((((...((((((....))))))((-(((.....)))))))).)))))))))))....((((((((........)))))))))))))))............ ( -44.00) >DroSec_CAF1 11521 119 - 1 AGUGCGCUCAAAAUGCGGCUCAGCUCAGCGGGAUUCCCUGUUUU-GCACUAUCUGCAACUGUGGCUGCUUUUGUAUCUGAGUGCCAUUGAAAAGGCGUUCAAGCGCGCAAUUUCGCAUGU .((((((((((((.(((((((((...((((((....))))))((-(((.....)))))))).)))))))))))....((((((((........)))))))))))))))............ ( -44.00) >DroSim_CAF1 10356 119 - 1 AGUGCGCUCAAAAUGCGGCUCAGCUCAGCGGGAUUCCCUGUUGU-GCACUAUCUGCAACUGUGGCUGCUUUUGUAUCUGAGUGCCAUUGAAGAGGCGUUCAAGCGCGCAAUUUCGCAUGU .((((((((((((.(((((((((..(((((((....)))))))(-(((.....)))).))).)))))))))))....((((((((........)))))))))))))))............ ( -46.30) >DroEre_CAF1 13279 120 - 1 AGUGCGCUCAAAAUGCGGCUCAGCUCAGCGGGAUUCCCUUUUUUCGCACUAUCUGCAACUGUGGCCGCUUUUGUAUCUGAGUGCCAUUGAAAAGGCGUUCAAGCGCGCAAUUUCGCAUGU .((((((((((((.(((((((((......(((...))).......(((.....)))..))).)))))))))))....((((((((........)))))))))))))))............ ( -41.70) >DroYak_CAF1 6637 119 - 1 AGUGCGCUCAAAAUGCGGCUCAGCUCAGCGGGAUUCCCUUUUUU-GCACUAUCUGCAACUGUGGCCGCUUUUGUAUCUGAGUGCCAUUGAAAAGGCGUUCAAUCGCGCAAUUUCGCAUGU ..(((((.(((((.(((((((((......(((...)))....((-(((.....)))))))).)))))))))))....((((((((........))))))))...)))))........... ( -39.20) >consensus AGUGCGCUCAAAAUGCGGCUCAGCUCAGCGGGAUUCCCUGUUUU_GCACUAUCUGCAACUGUGGCUGCUUUUGUAUCUGAGUGCCAUUGAAAAGGCGUUCAAGCGCGCAAUUUCGCAUGU .((((((((((((.(((((((((...((((((....))))))...(((.....)))..))).)))))))))))....((((((((........)))))))))))))))............ (-40.54 = -40.90 + 0.36)


| Location | 8,086,710 – 8,086,825 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.38 |
| Mean single sequence MFE | -35.44 |
| Consensus MFE | -31.88 |
| Energy contribution | -31.80 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8086710 115 - 27905053 AGAUUGGCAACAAUUGUGUGUGCAAUCGG-----CUCAUUGUGUUUUAACACUUUGAUGGGAAUCAGGGGUUGUUCGAGGUGGGCGUGUACUGACAAGUGACGACGAUUUUCCCACAUUA .(((((....)))))(((((...((((((-----(((((..((...((((.(((((((....)))))))))))..))..))))))((.((((....))))))..)))))....))))).. ( -35.90) >DroSec_CAF1 11640 115 - 1 AGAUUGGCAACAAUUGUGUGGGCAAUGGG-----CUCAUUGGGUUUUAACACUUUGAUGGGAAUCAGGGGUUGUUCGAGGUGGGCGUGUACUGACAAGUGACGACGAUUUUCCCACAUUA .(((((....)))))(((((((.(((.((-----((((((..(...((((.(((((((....)))))))))))..)..)))))))((.((((....))))))..).)))..))))))).. ( -38.00) >DroSim_CAF1 10475 115 - 1 AGAUUGGCAACAAUUGUGUGUGCAAUGGG-----CUCAUUGGGUUUUAACACUUUGAUGGGAAUCAGGGGUUGUUCGAGGUGGGCGUGUACUGACAAGUGACGACGAUUUUCCCACAUUA .(((((....)))))(((((...(((.((-----((((((..(...((((.(((((((....)))))))))))..)..)))))))((.((((....))))))..).)))....))))).. ( -31.00) >DroEre_CAF1 13399 120 - 1 AGAUUGGCAACAAUUGUGUGUGCAAUGGGCUCGGCUCAUUGUGCUCUAACACUUUGAUGGGAAUCAGGGGUUGUUCGAGGUGGGCGUGUACUGACAAGUGACGACGAUUUUCCCACAUUA .(((((....)))))..(.(..((((((((...))))))))..).)((((.(((((((....)))))))))))......(((((((((((((....))).)).))).....))))).... ( -38.80) >DroYak_CAF1 6756 115 - 1 AGAUCGGCAACAAUUGUGUGUGCUAUGGG-----CUCAUUGUGUUUUAACACUUUGAUGGGAAUCAGGGGUUGUUCGAGGUGGGCGUGCACUGACAAGUGACGACGAUUUUCCCACAUUA ((((((.(..((.((((..((((...(.(-----(((((..((...((((.(((((((....)))))))))))..))..)))))).)))))..)))).))..).)))))).......... ( -33.50) >consensus AGAUUGGCAACAAUUGUGUGUGCAAUGGG_____CUCAUUGUGUUUUAACACUUUGAUGGGAAUCAGGGGUUGUUCGAGGUGGGCGUGUACUGACAAGUGACGACGAUUUUCCCACAUUA ...((((((((...(((..(..(((((((.....)))))))..)....)))(((((((....))))))))))).)))).(((((((((((((....)))).).))).....))))).... (-31.88 = -31.80 + -0.08)



| Location | 8,086,750 – 8,086,865 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.87 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -21.74 |
| Energy contribution | -22.38 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8086750 115 + 27905053 CCUCGAACAACCCCUGAUUCCCAUCAAAGUGUUAAAACACAAUGAG-----CCGAUUGCACACACAAUUGUUGCCAAUCUGGAGAUACUCUCAUCUCCAGAAGCGGAUUGCCCAUGCUAA .((((.........((((....))))..((((....))))..))))-----......(((....(((((..(((...(((((((((......))))))))).))))))))....)))... ( -25.70) >DroSec_CAF1 11680 115 + 1 CCUCGAACAACCCCUGAUUCCCAUCAAAGUGUUAAAACCCAAUGAG-----CCCAUUGCCCACACAAUUGUUGCCAAUCUGGAGAUACUCUCAUCCCCAGAAGCGGAUUGCCCAUGCUAA ((...(((((....((((....))))..((((.......(((((..-----..)))))...))))..)))))((...(((((.(((......))).))))).)))).............. ( -19.20) >DroSim_CAF1 10515 115 + 1 CCUCGAACAACCCCUGAUUCCCAUCAAAGUGUUAAAACCCAAUGAG-----CCCAUUGCACACACAAUUGUUGCCAAUCUGGAGAUACUCUCAUCUCCAGAAGCGGAUUGCCCAUGCUAA ((...(((((....((((....))))..((((.......(((((..-----..)))))...))))..)))))((...(((((((((......))))))))).)))).............. ( -24.00) >DroEre_CAF1 13439 120 + 1 CCUCGAACAACCCCUGAUUCCCAUCAAAGUGUUAGAGCACAAUGAGCCGAGCCCAUUGCACACACAAUUGUUGCCAAUCUGGAGAUCCUCUCAUCUCCAGAAGCGGAUUGCCCAUGCUAA .((((.........((((....))))..((((....)))).......))))......(((....(((((..(((...(((((((((......))))))))).))))))))....)))... ( -27.90) >DroYak_CAF1 6796 115 + 1 CCUCGAACAACCCCUGAUUCCCAUCAAAGUGUUAAAACACAAUGAG-----CCCAUAGCACACACAAUUGUUGCCGAUCUGGAGAUCCUCUCAUCUCCAGAAGCGGAUUGCCCAUGCUAA .((((.........((((....))))..((((....))))..))))-----....(((((....(((((..(((...(((((((((......))))))))).))))))))....))))). ( -28.30) >consensus CCUCGAACAACCCCUGAUUCCCAUCAAAGUGUUAAAACACAAUGAG_____CCCAUUGCACACACAAUUGUUGCCAAUCUGGAGAUACUCUCAUCUCCAGAAGCGGAUUGCCCAUGCUAA .((((.........((((....))))..((((....))))..))))...........(((....(((((..(((...(((((((((......))))))))).))))))))....)))... (-21.74 = -22.38 + 0.64)

| Location | 8,086,750 – 8,086,865 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.87 |
| Mean single sequence MFE | -40.62 |
| Consensus MFE | -39.88 |
| Energy contribution | -39.96 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.98 |
| SVM decision value | 5.03 |
| SVM RNA-class probability | 0.999969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8086750 115 - 27905053 UUAGCAUGGGCAAUCCGCUUCUGGAGAUGAGAGUAUCUCCAGAUUGGCAACAAUUGUGUGUGCAAUCGG-----CUCAUUGUGUUUUAACACUUUGAUGGGAAUCAGGGGUUGUUCGAGG ......(((((((((((((((((((((((....))))))))))..)))..(.(((((....))))).).-----((((((((((....))))...))))))......))))))))))... ( -39.00) >DroSec_CAF1 11680 115 - 1 UUAGCAUGGGCAAUCCGCUUCUGGGGAUGAGAGUAUCUCCAGAUUGGCAACAAUUGUGUGGGCAAUGGG-----CUCAUUGGGUUUUAACACUUUGAUGGGAAUCAGGGGUUGUUCGAGG ......(((((((((((((((((((((((....))))))))))..)))...((((..((((((.....)-----)))))..))))........(((((....)))))))))))))))... ( -38.00) >DroSim_CAF1 10515 115 - 1 UUAGCAUGGGCAAUCCGCUUCUGGAGAUGAGAGUAUCUCCAGAUUGGCAACAAUUGUGUGUGCAAUGGG-----CUCAUUGGGUUUUAACACUUUGAUGGGAAUCAGGGGUUGUUCGAGG ......(((((((((((((((((((((((....))))))))))..))).......((((.(.(((((..-----..))))).).....)))).(((((....)))))))))))))))... ( -38.40) >DroEre_CAF1 13439 120 - 1 UUAGCAUGGGCAAUCCGCUUCUGGAGAUGAGAGGAUCUCCAGAUUGGCAACAAUUGUGUGUGCAAUGGGCUCGGCUCAUUGUGCUCUAACACUUUGAUGGGAAUCAGGGGUUGUUCGAGG ......((((((((((((((((((((((......)))))))))..))).......(((((..((((((((...))))))))..)....)))).(((((....)))))))))))))))... ( -47.80) >DroYak_CAF1 6796 115 - 1 UUAGCAUGGGCAAUCCGCUUCUGGAGAUGAGAGGAUCUCCAGAUCGGCAACAAUUGUGUGUGCUAUGGG-----CUCAUUGUGUUUUAACACUUUGAUGGGAAUCAGGGGUUGUUCGAGG .(((((((.((((((((..(((((((((......))))))))).))).....))))).)))))))....-----(((..(((......)))(((((((....))))))).......))). ( -39.90) >consensus UUAGCAUGGGCAAUCCGCUUCUGGAGAUGAGAGUAUCUCCAGAUUGGCAACAAUUGUGUGUGCAAUGGG_____CUCAUUGUGUUUUAACACUUUGAUGGGAAUCAGGGGUUGUUCGAGG ......((((((((((((((((((((((......)))))))))..))).......(((((..(((((((.....)))))))..)....)))).(((((....)))))))))))))))... (-39.88 = -39.96 + 0.08)


| Location | 8,086,790 – 8,086,905 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.73 |
| Mean single sequence MFE | -38.64 |
| Consensus MFE | -33.18 |
| Energy contribution | -33.42 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8086790 115 - 27905053 CUGGCACUUGAGUGUUCAUUGAGUUUUACGCAUCUGACGAUUAGCAUGGGCAAUCCGCUUCUGGAGAUGAGAGUAUCUCCAGAUUGGCAACAAUUGUGUGUGCAAUCGG-----CUCAUU ..(((((....)))))...(((((....((....)).(((((.(((((.(((((..(((((((((((((....))))))))))..)))....))))).)))))))))))-----)))).. ( -37.70) >DroSec_CAF1 11720 115 - 1 CUGGCACUUGAGUGUUCAUUGAGUUUUACGCAUCUGACGAUUAGCAUGGGCAAUCCGCUUCUGGGGAUGAGAGUAUCUCCAGAUUGGCAACAAUUGUGUGGGCAAUGGG-----CUCAUU ........(((((..((((((....(((((((..((..((((.((....)))))).(((((((((((((....))))))))))..)))..))..))))))).)))))))-----)))).. ( -37.10) >DroSim_CAF1 10555 115 - 1 CGGGCACUUGAGUGUUCAUUGAGUUUUACGCAUCUGACGAUUAGCAUGGGCAAUCCGCUUCUGGAGAUGAGAGUAUCUCCAGAUUGGCAACAAUUGUGUGUGCAAUGGG-----CUCAUU .((((((....))))))..(((((((...((((...((((((.((....)).....(((((((((((((....))))))))))..)))...))))))..))))...)))-----)))).. ( -39.70) >DroEre_CAF1 13479 120 - 1 CUGGCACUUGAGUGUUCAUUGAGUUUUACGCAUCUGACGAUUAGCAUGGGCAAUCCGCUUCUGGAGAUGAGAGGAUCUCCAGAUUGGCAACAAUUGUGUGUGCAAUGGGCUCGGCUCAUU ........(((((((((((((.....((((((..((..((((.((....)))))).((((((((((((......)))))))))..)))..))..))))))..))))))))...))))).. ( -40.50) >DroYak_CAF1 6836 115 - 1 CUGGCACUUGAGUGUUCAUUGAGUUUUACGCAUCUGACGAUUAGCAUGGGCAAUCCGCUUCUGGAGAUGAGAGGAUCUCCAGAUCGGCAACAAUUGUGUGUGCUAUGGG-----CUCAUU ..(((((....)))))...(((((((..(((....).))..(((((((.((((((((..(((((((((......))))))))).))).....))))).))))))).)))-----)))).. ( -38.20) >consensus CUGGCACUUGAGUGUUCAUUGAGUUUUACGCAUCUGACGAUUAGCAUGGGCAAUCCGCUUCUGGAGAUGAGAGUAUCUCCAGAUUGGCAACAAUUGUGUGUGCAAUGGG_____CUCAUU ........((((..(((((((.....((((((..((..((((.((....)))))).((((((((((((......)))))))))..)))..))..))))))..))))))).....)))).. (-33.18 = -33.42 + 0.24)



| Location | 8,086,905 – 8,087,023 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.48 |
| Mean single sequence MFE | -30.82 |
| Consensus MFE | -20.58 |
| Energy contribution | -20.57 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8086905 118 - 27905053 CCCGGUUUCUCAUGAUCGGAUUAAAAGGAUUAUGAGUUCGCGAUCUCGGUAGUCUGUCUGUUUGUUUUU--UUAUUUUUCAGUAGCCUCUUAGCUGCUUUGGGACUUUUGUCCCGGAACC ...((((((....((.(((((((...((((..((....))..))))...)))))))))...........--.........((((((......))))))..(((((....))))))))))) ( -34.60) >DroSec_CAF1 11835 107 - 1 CCCGGUUUCUCAUGAUCGGAUUGAAAGGAUUAUGAGUUCGCGAUCUUGGUAGUCUGGCUGUUUGUUUUUUUUUCUUUUUCAGUAGCCUCUUAGCUGCUUUGGGAC-------------CC ...(((..(......((((((((..(((((..((....))..)))))..)))))))).......................((((((......))))))..)..))-------------). ( -27.90) >DroEre_CAF1 13599 116 - 1 CGCGUUUUCUCAUGAUCGGAUUGAAAGGAUUAUGAGUUCGCGAUCUCGGUUGUCUGUCUGUUUGUUUU---UUAUUGUUCAGUAGCUUCUUAGCUGCUUUGGGACUUCUGC-CAGGACCC ((((....(((((((((..........)))))))))..)))).((..(((.(...((((...((..(.---.....)..))((((((....))))))....))))..).))-)..))... ( -32.30) >DroYak_CAF1 6951 116 - 1 CCCGUUUUCUCAUGAUCGGAUUGGAAGGAUUAUGAGUUCGCGAUCUUGGUAGUCGCUCUGUUUGUUUU---UUAUUUUUCAGUAGCUUCUUAGCUGCUUUGGGACUUUUGU-CAGGACCC ..(((...(((((((((..........)))))))))...))).((((((.((((((.......))...---.........(((((((....)))))))....))))....)-)))))... ( -28.50) >consensus CCCGGUUUCUCAUGAUCGGAUUGAAAGGAUUAUGAGUUCGCGAUCUCGGUAGUCUGUCUGUUUGUUUU___UUAUUUUUCAGUAGCCUCUUAGCUGCUUUGGGACUUUUGU_CAGGACCC ((((....(((((((((..........)))))))))...((((...(((........))).))))...............((((((......)))))).))))................. (-20.58 = -20.57 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:12 2006