
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,085,670 – 8,086,164 |
| Length | 494 |
| Max. P | 0.999978 |

| Location | 8,085,670 – 8,085,782 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.49 |
| Mean single sequence MFE | -40.96 |
| Consensus MFE | -38.02 |
| Energy contribution | -39.18 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.93 |
| SVM decision value | 5.20 |
| SVM RNA-class probability | 0.999978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8085670 112 - 27905053 CCAUUCGUAUCUCACAGUUGCAUCUUGUUAUCUCGUCAAGAGAUUCGAGUGCGAGUGUGAGUGUGCGUGGGUAAACUCUCUGCUUGGCGACAAUUUUGCGUCAUUUGCGGUU-------- (((..(((((((((((.(((((.((((..(((((.....))))).))))))))).)))))).)))))))).........((((.(((((.((....)))))))...))))..-------- ( -44.20) >DroSec_CAF1 10592 112 - 1 CCAUUCGUAUCUCACAGUUGCAUCUUGUUAUCUCGUCAAGAGAUUCGAGUGCGAGUGUGAGUGUGCGUGGGUAAACUCUCUGCUUGGCGACAAUUUUGCGUCAUUUGCGGUU-------- (((..(((((((((((.(((((.((((..(((((.....))))).))))))))).)))))).)))))))).........((((.(((((.((....)))))))...))))..-------- ( -44.20) >DroSim_CAF1 9428 112 - 1 CCAUUCGUAUCUCACAGUUGCAUCUUGUUAUCUCGUCAAGAGAUUCGAGUGCGAGUGUGAGUGUGCGUGGGUAAACUCUCUGCUUGGCGACAAUUUUGCGUCAUUUGCGGUU-------- (((..(((((((((((.(((((.((((..(((((.....))))).))))))))).)))))).)))))))).........((((.(((((.((....)))))))...))))..-------- ( -44.20) >DroEre_CAF1 12349 106 - 1 CCAGUCGUAUCUCACAGUUGCAUCUUGUUAUCUCGUCAAGAGAUUCGAGUGUGAGUGU------GCGUGGGUAAACUCUCUGCUUGGCGACAAUUUUGCGUCAUUUGCGGUU-------- (((..(((((((((((.(((..(((((.........)))))....))).)))))).))------)))))).........((((.(((((.((....)))))))...))))..-------- ( -32.90) >DroYak_CAF1 5702 120 - 1 CCAGUCGUAUCUCACAGUUGCAUCUUUUUAUCUCGUCAAGAGAUUCGAGUGUGAGUGUGCGUGUGCGUGGGUAAACUCUCUGCUUGGCGACAAUUUUGCGUCAUUUGCGGUUGGUUGUCU (((..((((((.((((.(..((.(((...(((((.....)))))..)))))..).)))).).))))))))...((((..((((.(((((.((....)))))))...))))..)))).... ( -39.30) >consensus CCAUUCGUAUCUCACAGUUGCAUCUUGUUAUCUCGUCAAGAGAUUCGAGUGCGAGUGUGAGUGUGCGUGGGUAAACUCUCUGCUUGGCGACAAUUUUGCGUCAUUUGCGGUU________ (((..(((((((((((.(((((.((((..(((((.....))))).))))))))).)))))).)))))))).........((((.(((((.((....)))))))...)))).......... (-38.02 = -39.18 + 1.16)

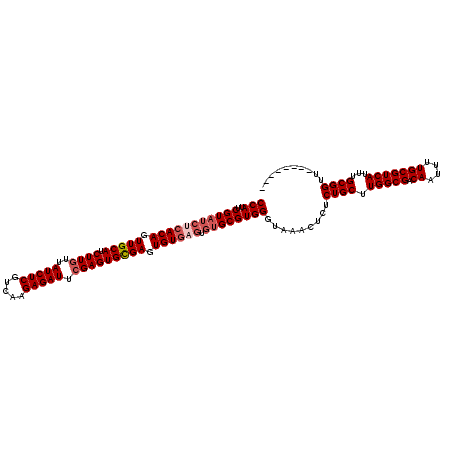

| Location | 8,085,702 – 8,085,822 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.75 |
| Mean single sequence MFE | -42.32 |
| Consensus MFE | -38.60 |
| Energy contribution | -39.76 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.89 |
| SVM RNA-class probability | 0.999685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8085702 120 - 27905053 AAGCUGUGUGCGAGGGCCUCAUUAAACGGUAUUUUUGUGGCCAUUCGUAUCUCACAGUUGCAUCUUGUUAUCUCGUCAAGAGAUUCGAGUGCGAGUGUGAGUGUGCGUGGGUAAACUCUC ..((.....))(((((((.((..((......))..)).))))...(((((((((((.(((((.((((..(((((.....))))).))))))))).)))))).)))))........))).. ( -45.00) >DroSec_CAF1 10624 120 - 1 AAGCUGUGUGCGAGGGCCUCAUUAAACGGUAUUUUUGUGGCCAUUCGUAUCUCACAGUUGCAUCUUGUUAUCUCGUCAAGAGAUUCGAGUGCGAGUGUGAGUGUGCGUGGGUAAACUCUC ..((.....))(((((((.((..((......))..)).))))...(((((((((((.(((((.((((..(((((.....))))).))))))))).)))))).)))))........))).. ( -45.00) >DroSim_CAF1 9460 120 - 1 AAGCUGUGUGCGAGGGCCUCAUUAAACGGUAUUUUUGUGGCCAUUCGUAUCUCACAGUUGCAUCUUGUUAUCUCGUCAAGAGAUUCGAGUGCGAGUGUGAGUGUGCGUGGGUAAACUCUC ..((.....))(((((((.((..((......))..)).))))...(((((((((((.(((((.((((..(((((.....))))).))))))))).)))))).)))))........))).. ( -45.00) >DroEre_CAF1 12381 114 - 1 AAGCUGUGUGCGAGGGCCUCAUUAAACGGUAUUUUUGUGGCCAGUCGUAUCUCACAGUUGCAUCUUGUUAUCUCGUCAAGAGAUUCGAGUGUGAGUGU------GCGUGGGUAAACUCUC .((((((((((((.((((.((..((......))..)).))))..)))))...)))))))(((.((((..(((((.....))))).)))))))((((.(------((....))).)))).. ( -39.00) >DroYak_CAF1 5742 120 - 1 AAGCUGUGAGCGAGGGCCUCAUUAAACGGUAUUUCUGUGGCCAGUCGUAUCUCACAGUUGCAUCUUUUUAUCUCGUCAAGAGAUUCGAGUGUGAGUGUGCGUGUGCGUGGGUAAACUCUC .((((((((((((.((((.((..((......))..)).))))..)))...)))))))))(((.(((...(((((.....)))))..))))))((((.(((.(......).))).)))).. ( -37.60) >consensus AAGCUGUGUGCGAGGGCCUCAUUAAACGGUAUUUUUGUGGCCAUUCGUAUCUCACAGUUGCAUCUUGUUAUCUCGUCAAGAGAUUCGAGUGCGAGUGUGAGUGUGCGUGGGUAAACUCUC ..((.....))(((((((.((..((......))..)).))))...(((((((((((.(((((.((((..(((((.....))))).))))))))).)))))).)))))........))).. (-38.60 = -39.76 + 1.16)



| Location | 8,085,742 – 8,085,857 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.59 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -24.82 |
| Energy contribution | -24.82 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8085742 115 + 27905053 CUUGACGAGAUAACAAGAUGCAACUGUGAGAUACGAAUGGCCACAAAAAUACCGUUUAAUGAGGCCCUCGCACACAGCUUGACUAUUUCUUAGAUCUUAU-----CCGUAGGGAAGUCAC ..(((((((((...((((.(((((((((.(...(((..((((.((..((......))..)).)))).)))).))))).))).)....))))..))))).(-----((....))).)))). ( -27.30) >DroSec_CAF1 10664 119 + 1 CUUGACGAGAUAACAAGAUGCAACUGUGAGAUACGAAUGGCCACAAAAAUACCGUUUAAUGAGGCCCUCGCACACAGCUUGACUAUUUCUUAGAUCUUAUCCCAUCCGUAGGG-AGUCAC ..(((((((((...((((.(((((((((.(...(((..((((.((..((......))..)).)))).)))).))))).))).)....))))..))))).((((.......)))-))))). ( -30.20) >DroSim_CAF1 9500 119 + 1 CUUGACGAGAUAACAAGAUGCAACUGUGAGAUACGAAUGGCCACAAAAAUACCGUUUAAUGAGGCCCUCGCACACAGCUUGACUAUUUCUUAGAUCUUAUCCCAUCCGUAGGG-AGUCAC ..(((((((((...((((.(((((((((.(...(((..((((.((..((......))..)).)))).)))).))))).))).)....))))..))))).((((.......)))-))))). ( -30.20) >DroEre_CAF1 12415 114 + 1 CUUGACGAGAUAACAAGAUGCAACUGUGAGAUACGACUGGCCACAAAAAUACCGUUUAAUGAGGCCCUCGCACACAGCUUGACUAUUUCUUAGAUCUUAU-----CCGUAGGG-AGUCAC ..(((((((((...((((.(((((((((.(...(((..((((.((..((......))..)).)))).)))).))))).))).)....))))..))))).(-----((....))-))))). ( -27.30) >DroYak_CAF1 5782 114 + 1 CUUGACGAGAUAAAAAGAUGCAACUGUGAGAUACGACUGGCCACAGAAAUACCGUUUAAUGAGGCCCUCGCUCACAGCUUGACUAUUUCUUAGAUCUUAU-----CCGUAGGG-AGUCAC ..(((((((((...((((.(((((((((((...(((..((((.((..((......))..)).)))).)))))))))).))).)....))))..))))).(-----((....))-))))). ( -33.50) >consensus CUUGACGAGAUAACAAGAUGCAACUGUGAGAUACGAAUGGCCACAAAAAUACCGUUUAAUGAGGCCCUCGCACACAGCUUGACUAUUUCUUAGAUCUUAU_____CCGUAGGG_AGUCAC ..(((((((((...((((.(((((((((.(...(((..((((.((..((......))..)).)))).)))).))))).))).)....))))..))))).......((....))..)))). (-24.82 = -24.82 + 0.00)



| Location | 8,085,857 – 8,085,968 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.29 |
| Mean single sequence MFE | -30.41 |
| Consensus MFE | -23.76 |
| Energy contribution | -24.16 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8085857 111 + 27905053 UCAAAGGAUAUUGGC-CGGCUGCCUAUUCUCUAUGAAAUCGAUUCUAUUCAUCCCGCU--------UUUUCGGGUUUCCGAGGCCCAUAACUCGUGUUACUUUAUGCAGCCGGCUUUGUU ......((((..(((-(((((((...........((((..((......))..((((..--------....))))))))((((........))))...........)))))))))).)))) ( -32.80) >DroSec_CAF1 10783 112 + 1 UCAAAGGAUAUUGGCCCGGCUGCCUAUUCUCAUUGAAAUCGAUUCUAUUCAUCCCGCU--------UUUUCGGGUUUCCGAGGCCCAUAACUCGUGUUACUUUAUGCAGCCGGCUUUGUU ......((((..((((.((((((...........((((..((......))..((((..--------....))))))))((((........))))...........)))))))))).)))) ( -29.00) >DroSim_CAF1 9619 111 + 1 UCAAAGGAUAUUGGCCCGGCUGCCUAUUCUCAUUGAAAUCGAUUCUAUUCAUC-CGCU--------UUUUCGGGUUUCCGAGGCCCAUAACUCGUGUUGCUUUAUGCAGCCGGCUUUGUU .(((((..(((.(((((((..((((.(((.....)))...((......))...-....--------.....))))..))).)))).)))..(((.(((((.....))))))))))))).. ( -32.30) >DroEre_CAF1 12529 118 + 1 UCAAAGGAUAUUGGC-CGGCUGCCUAUUCUCAUUGAAAUCGAAUCUAUUCAUCC-ACUAUCCACACUAUUCGGGUUUCCGAGGCCCAUAACUCCUGGUACUUUAUGCAGCCGGCUUUGUU ......((((..(((-(((((((...........((((((((((..........-............)))).)))))).(((..(((.......)))..)))...)))))))))).)))) ( -30.05) >DroYak_CAF1 5896 116 + 1 UCAAAGGAUAUUGGC-CGGCUGCCUAUUCUCAUUGAAAUCGAUUCUAUUCAGCCUACUA---CUACUAUUCGUGUUUCCGAUGCCCAUAACUCCUGGUACUUUAUGCAGCCGGCUUUGUU ......((((..(((-(((((((.......((((((((((((........((....)).---.......))).)))).))))).(((.......)))........)))))))))).)))) ( -27.89) >consensus UCAAAGGAUAUUGGC_CGGCUGCCUAUUCUCAUUGAAAUCGAUUCUAUUCAUCCCGCU________UUUUCGGGUUUCCGAGGCCCAUAACUCGUGUUACUUUAUGCAGCCGGCUUUGUU ......((((..(((.(((((((...(((.....)))..................................(((((.....)))))...................)))))))))).)))) (-23.76 = -24.16 + 0.40)



| Location | 8,085,968 – 8,086,088 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.42 |
| Mean single sequence MFE | -28.24 |
| Consensus MFE | -27.50 |
| Energy contribution | -27.18 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8085968 120 + 27905053 UGACCACAAAGUGUCGCUGACCAAGUCCCCAUAAUCGAUUGAAUUACUUGCUCCCUAGCAGCUUCCUUCCGCAAUUCAUGUCAGGCUCUAAAUUCAAAGGUUUAGAGCCCCAAAAUGUGC ....((((..(((..(((.....)))...)))....(((((((((...((((....))))((........)))))))).))).((((((((((......))))))))))......)))). ( -27.60) >DroSec_CAF1 10895 120 + 1 UGACCACAAAGUGUCGCUGACCAAGUCCCCAUAAUCGAUUGAAUUACCCGCUCCCUAGCAGCUUCCUUCCGCAAUUCUUGUCAGGCUCUAAAUUCAAGGGUUUAGAGCCCCAAAAUGUGC ....((((..(((..(((.....)))...)))....(((.(((((....(((....))).((........)))))))..))).(((((((((((....)))))))))))......)))). ( -27.50) >DroSim_CAF1 9730 120 + 1 UGACCACAAAGUGUCGCUGACCAAGUCUCCAUAAUCGAUUGAAUUACCCGCUCCCUAGCAGCUUCCUUCCGCAAUUCAUGUCAGGCUCUAAAUUCAAGGGUUUAGAGCCCCAAAAUGUGC ....((((..(((..(((.....)))...)))....(((((((((....(((....))).((........)))))))).))).(((((((((((....)))))))))))......)))). ( -28.40) >DroEre_CAF1 12647 120 + 1 UGACCACAAAGUGUCGCUGACCAAGUCCCUAUAAUCGAUUGAAUUACCCGCUCCCAAGCAGCUUCCUUCCGCAAUUCCUGUCAGGCUCUAAAUUCAAGGGUUUAGAGCCCCAAAAUGUGC ....((((....(..(((.....)))..).......(((.(((((....(((....))).((........)))))))..))).(((((((((((....)))))))))))......)))). ( -26.90) >DroYak_CAF1 6012 120 + 1 UGACCACAAAGUGUCGCUGACCAAGUCCCCAUAAUCGAUUGAAUUACCCGCUCUCUGGCAGCUUCCUUCCGCAAUUCAUGUCAGGCUCUAAACUCAAGGGUUUAGAGCCCCAAAAUGUGC ....((((..(((..(((.....)))...)))....(((((((((....(((....))).((........)))))))).))).(((((((((((....)))))))))))......)))). ( -30.80) >consensus UGACCACAAAGUGUCGCUGACCAAGUCCCCAUAAUCGAUUGAAUUACCCGCUCCCUAGCAGCUUCCUUCCGCAAUUCAUGUCAGGCUCUAAAUUCAAGGGUUUAGAGCCCCAAAAUGUGC ....((((...((....((((..((((.........))))(((((....(((....))).((........)))))))..))))(((((((((((....))))))))))).))...)))). (-27.50 = -27.18 + -0.32)

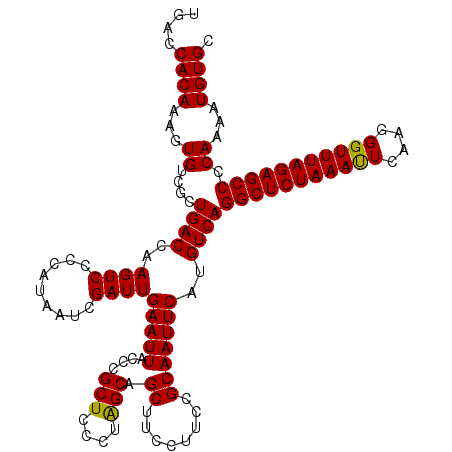
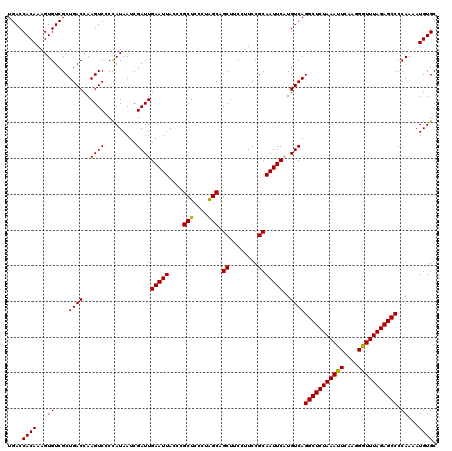
| Location | 8,086,008 – 8,086,128 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.42 |
| Mean single sequence MFE | -33.44 |
| Consensus MFE | -27.56 |
| Energy contribution | -28.24 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8086008 120 + 27905053 GAAUUACUUGCUCCCUAGCAGCUUCCUUCCGCAAUUCAUGUCAGGCUCUAAAUUCAAAGGUUUAGAGCCCCAAAAUGUGCAGCGUCAACUGUCGAGAAUGGAUUUAGUGGCAGUGCAUCU ........((((....))))((...((.(((((((((((.((.((((((((((......)))))))))).........((((......)))).))..)))))))..)))).)).)).... ( -32.40) >DroSec_CAF1 10935 120 + 1 GAAUUACCCGCUCCCUAGCAGCUUCCUUCCGCAAUUCUUGUCAGGCUCUAAAUUCAAGGGUUUAGAGCCCCAAAAUGUGCAGCGUCAACUGUCGAGAAAGGAUUUAGUGGCAGUGCAUCU .........(((....))).((...((.(((((((((((.((.(((((((((((....))))))))))).........((((......)))).))..)))))))..)))).)).)).... ( -33.00) >DroSim_CAF1 9770 120 + 1 GAAUUACCCGCUCCCUAGCAGCUUCCUUCCGCAAUUCAUGUCAGGCUCUAAAUUCAAGGGUUUAGAGCCCCAAAAUGUGCAGCGUCAACUGUCGAGAAAGGAUUUAGUGGCAGUGCAUCU ........((((((((((.....(((((.((((...(((....(((((((((((....))))))))))).....))))))..((.(....).)).).))))).)))).)).))))..... ( -31.80) >DroEre_CAF1 12687 120 + 1 GAAUUACCCGCUCCCAAGCAGCUUCCUUCCGCAAUUCCUGUCAGGCUCUAAAUUCAAGGGUUUAGAGCCCCAAAAUGUGCAGCGUCAACUGUCGAUAAAGGAUUUAAAGGAACUGCAUCC .................((((.((((((.......(((((((.(((((((((((....))))))))))).........((((......)))).)))..))))....)))))))))).... ( -35.50) >DroYak_CAF1 6052 120 + 1 GAAUUACCCGCUCUCUGGCAGCUUCCUUCCGCAAUUCAUGUCAGGCUCUAAACUCAAGGGUUUAGAGCCCCAAAAUGUGCAGCGUCAACUGUCGAGAAAGGAUUUUAAAGAACUGCAUCU .......((..((((.(((((........(((....(((....(((((((((((....))))))))))).....)))....)))....)))))))))..))................... ( -34.50) >consensus GAAUUACCCGCUCCCUAGCAGCUUCCUUCCGCAAUUCAUGUCAGGCUCUAAAUUCAAGGGUUUAGAGCCCCAAAAUGUGCAGCGUCAACUGUCGAGAAAGGAUUUAGUGGCAGUGCAUCU (((((....(((....))).((........)))))))......(((((((((((....))))))))))).......(((((..((((...(((.......)))....))))..))))).. (-27.56 = -28.24 + 0.68)



| Location | 8,086,008 – 8,086,128 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.42 |
| Mean single sequence MFE | -36.38 |
| Consensus MFE | -33.92 |
| Energy contribution | -34.36 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8086008 120 - 27905053 AGAUGCACUGCCACUAAAUCCAUUCUCGACAGUUGACGCUGCACAUUUUGGGGCUCUAAACCUUUGAAUUUAGAGCCUGACAUGAAUUGCGGAAGGAAGCUGCUAGGGAGCAAGUAAUUC ...(((..(((............((((..(((((..(.(((((.(((((.((((((((((........)))))))))).)...)))))))))..)..)))))...))))))).))).... ( -30.90) >DroSec_CAF1 10935 120 - 1 AGAUGCACUGCCACUAAAUCCUUUCUCGACAGUUGACGCUGCACAUUUUGGGGCUCUAAACCCUUGAAUUUAGAGCCUGACAAGAAUUGCGGAAGGAAGCUGCUAGGGAGCGGGUAAUUC .................((((.(((((..(((((..(.(((((..(((((((((((((((........))))))))))..)))))..)))))..)..)))))...))))).))))..... ( -38.00) >DroSim_CAF1 9770 120 - 1 AGAUGCACUGCCACUAAAUCCUUUCUCGACAGUUGACGCUGCACAUUUUGGGGCUCUAAACCCUUGAAUUUAGAGCCUGACAUGAAUUGCGGAAGGAAGCUGCUAGGGAGCGGGUAAUUC .................((((.(((((..(((((..(.(((((.(((((.((((((((((........)))))))))).)...)))))))))..)..)))))...))))).))))..... ( -36.10) >DroEre_CAF1 12687 120 - 1 GGAUGCAGUUCCUUUAAAUCCUUUAUCGACAGUUGACGCUGCACAUUUUGGGGCUCUAAACCCUUGAAUUUAGAGCCUGACAGGAAUUGCGGAAGGAAGCUGCUUGGGAGCGGGUAAUUC (((......))).....((((....(((((((((..(.(((((..(((((((((((((((........))))))))))..)))))..)))))..)..))))).))))....))))..... ( -36.60) >DroYak_CAF1 6052 120 - 1 AGAUGCAGUUCUUUAAAAUCCUUUCUCGACAGUUGACGCUGCACAUUUUGGGGCUCUAAACCCUUGAGUUUAGAGCCUGACAUGAAUUGCGGAAGGAAGCUGCCAGAGAGCGGGUAAUUC .................((((.(((((..(((((..(.(((((.(((((.(((((((((((......))))))))))).)...)))))))))..)..)))))...))))).))))..... ( -40.30) >consensus AGAUGCACUGCCACUAAAUCCUUUCUCGACAGUUGACGCUGCACAUUUUGGGGCUCUAAACCCUUGAAUUUAGAGCCUGACAUGAAUUGCGGAAGGAAGCUGCUAGGGAGCGGGUAAUUC .................((((.(((((..(((((..(.(((((.(((((.((((((((((........)))))))))).)...)))))))))..)..)))))...))))).))))..... (-33.92 = -34.36 + 0.44)



| Location | 8,086,048 – 8,086,164 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.18 |
| Mean single sequence MFE | -38.00 |
| Consensus MFE | -28.00 |
| Energy contribution | -28.04 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8086048 116 + 27905053 UCAGGCUCUAAAUUCAAAGGUUUAGAGCCCCAAAAUGUGCAGCGUCAACUGUCGAGAAUGGAUUUAGUGGCAGUGCAUCUUAGCAGGAA---ACUACUACGGAUUGGUUGG-AUGCUGGC ...((((((((((......))))))))))..........(((((((.(((((((..((.....))..)))))))..(((((((.((...---.)).))).))))......)-)))))).. ( -40.10) >DroSec_CAF1 10975 116 + 1 UCAGGCUCUAAAUUCAAGGGUUUAGAGCCCCAAAAUGUGCAGCGUCAACUGUCGAGAAAGGAUUUAGUGGCAGUGCAUCUUAGCAGGAA---ACUAGUACCGAUUGGUUGG-AUGCUGGC ...(((((((((((....)))))))))))..........(((((((.(((((((..((.....))..)))))))((......))....(---((((((....))))))).)-)))))).. ( -38.70) >DroSim_CAF1 9810 116 + 1 UCAGGCUCUAAAUUCAAGGGUUUAGAGCCCCAAAAUGUGCAGCGUCAACUGUCGAGAAAGGAUUUAGUGGCAGUGCAUCUUAGCAGGAA---ACUACUACCAAUUGGUUGG-AUGCUGGC ...(((((((((((....)))))))))))..........(((((((.(((((((..((.....))..)))))))((......))((...---.)).(.(((....))).))-)))))).. ( -39.40) >DroEre_CAF1 12727 116 + 1 UCAGGCUCUAAAUUCAAGGGUUUAGAGCCCCAAAAUGUGCAGCGUCAACUGUCGAUAAAGGAUUUAAAGGAACUGCAUCCUAGCUGGAA---ACUACUACUGAUUGGUUGG-AUGCUGGC ...(((((((((((....)))))))))))..........(((((((((((((((....(((((.((.......)).)))))((..(...---.)..))..))).))))).)-)))))).. ( -35.40) >DroYak_CAF1 6092 120 + 1 UCAGGCUCUAAACUCAAGGGUUUAGAGCCCCAAAAUGUGCAGCGUCAACUGUCGAGAAAGGAUUUUAAAGAACUGCAUCUUAGCAGGAUGCUACUACUACUGAUUGGUUGGAAUGCUGGC ...(((((((((((....)))))))))))..........((((((((((((((.((..((............))((((((.....))))))........)))).))))))..)))))).. ( -36.40) >consensus UCAGGCUCUAAAUUCAAGGGUUUAGAGCCCCAAAAUGUGCAGCGUCAACUGUCGAGAAAGGAUUUAGUGGCAGUGCAUCUUAGCAGGAA___ACUACUACCGAUUGGUUGG_AUGCUGGC ...(((((((((((....)))))))))))..........((((((((((((((.......)))..........(((......)))....................)))))..)))))).. (-28.00 = -28.04 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:02 2006