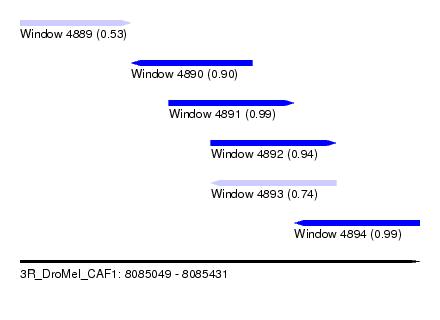
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,085,049 – 8,085,431 |
| Length | 382 |
| Max. P | 0.989125 |

| Location | 8,085,049 – 8,085,155 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.27 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -18.94 |
| Energy contribution | -19.54 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.533140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8085049 106 + 27905053 AAAUACAUAAAUA------UAUACUCUACAGAUGUCGCAUCUUAUCA--------GUCCGCUGAGGACCCAAAGUGAUGUCCACAUCAGGCGAGAGCUGUGAGAUUAAUUCUACACAUAG .............------....(((.((((.(.((((.........--------((((.....))))......(((((....))))).)))).).)))))))................. ( -25.40) >DroSec_CAF1 9976 106 + 1 AAAUACACAAAUA------UAUACUCUACAGAUUUCGCAUCUUAUCA--------GUCCGCUGAGGACCCAAAGUGAUGUCCACAUCAGGCGAGAGUUGUGAGAUUAAUUCUGCACAUAG .............------....(((.((((.((((((.........--------((((.....))))......(((((....))))).)))))).)))))))................. ( -24.80) >DroSim_CAF1 8811 106 + 1 AAAUACAUAAAUA------UAUACUCUACAGAUGUCGCAUCUUAUCA--------GUCCGCUGAGGACCCAAAGUGAUGUCCACAUCAGGCGAGAGUUGUGAGAUUAAUUCUGCACAUAG .............------...(((((.(.(((((.(((((......--------((((.....)))).......)))).).))))).)...)))))(((((((.....))).))))... ( -25.42) >DroEre_CAF1 11732 115 + 1 AAAUACAUAAAUAUACGAGUAUACUAUACAAAUGUCGCAUCUUAUCAG-UCGUCAGUCCGCUGAGGACCCAAUGAGAUGUCCACAUCAGGCGAGAGUUGGGAGAUUAAUUCUACAC---- ...............((.((((...))))..((((.(((((((((..(-((.((((....)))).)))...)))))))).).))))....))((((((((....))))))))....---- ( -26.50) >DroYak_CAF1 5085 116 + 1 AAAUACAUAAAUACUCGCAUAUACUGUACAAAUGUCGCAUCUUAUCAGCUCGUCGGUCCGCUGAGGACCCAAAGUGAUGUCCACAUCAGGCGAGAGUUUUGAGAUUAAUUCUACAC---- ................((((((.........)))).))((((((....(((((((((((.....))))).....(((((....))))))))))).....))))))...........---- ( -28.90) >consensus AAAUACAUAAAUA______UAUACUCUACAGAUGUCGCAUCUUAUCA________GUCCGCUGAGGACCCAAAGUGAUGUCCACAUCAGGCGAGAGUUGUGAGAUUAAUUCUACACAUAG .......................(((.((((.(.((((.................((((.....))))......(((((....))))).)))).).)))))))................. (-18.94 = -19.54 + 0.60)

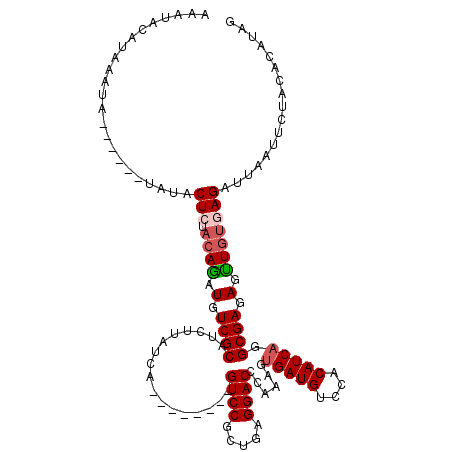
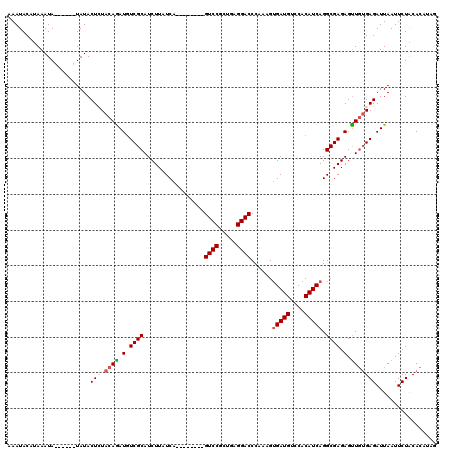
| Location | 8,085,155 – 8,085,271 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.88 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -32.56 |
| Energy contribution | -31.68 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8085155 116 - 27905053 CGCGGUUCAACGUGAACCUAAUUGAAAUUGUUCACCCUAAAAAUUGAUGUCGCCAAAGAUCAGGGAAUGCGGCUGUUCGUCGCAGGACAAAAGUUCAGUGG----GCAGCCAUGUAUGUA .(((((((.....)))))..((((((.((((((.((((.....(((.......))).....))))..((((((.....))))))))))))...))))))..----))............. ( -32.50) >DroSec_CAF1 10082 116 - 1 CGCGGUUCAACGUGAACCUAAUUGAAAUUGUUCACCCUAAAAAUUGAUGUCGCCAAAGAUCAGGGAACGCGGCUGUUCGUCGCAGGACAAAAGUUCAGUGG----GUAGCCAUGUAUGUA ((((......)))).(((((.(((((.((((((.((((.....(((.......))).....))))...(((((.....))))).))))))...))))))))----))............. ( -32.00) >DroSim_CAF1 8917 120 - 1 CGCGGUUCAACGUGAACCUAAUUGAAAUUGUUCACCCUAAAAAUUGAUGUCGCCAAAGAUCAGGGAACGCGGCUGUUCGUCGCAGGACAAAAGUUCAGUGGGCGGGCAGCCAUGUAUGUA .(((((((.....)))))...........((((.((........((((.(......).))))))))))))(((((((((((((.((((....)))).)).)))))))))))......... ( -38.60) >DroEre_CAF1 11847 113 - 1 CGCGGUUCAACGUGAACCUAAUUGAAAUUGUUCACCCUAAAAAUUGAUGUCGCCAAAGAUCAGGGAACGCGGCUGUUCGUCGCAGGACAAGAGUUCAGUGG----ACAGCCAUGUAU--- .(((((((.....)))))..((((((.((((((.((((.....(((.......))).....))))...(((((.....))))).))))))...))))))..----...)).......--- ( -32.20) >DroYak_CAF1 5201 113 - 1 CGCGGUUCAACGUGAACCUAAUUGAAAUUGUUCACCCUAAAAAUUGAUGUCGCCAAAGAUCAGGGAACGCGGCUGUUCGCCGCAGGACAAAAGUUCAGCGG----ACAGCCAUGUAU--- .(((((((.....)))))...........((((.((........((((.(......).))))))))))))((((((((((....((((....)))).))))----))))))......--- ( -36.20) >consensus CGCGGUUCAACGUGAACCUAAUUGAAAUUGUUCACCCUAAAAAUUGAUGUCGCCAAAGAUCAGGGAACGCGGCUGUUCGUCGCAGGACAAAAGUUCAGUGG____GCAGCCAUGUAUGUA .(((((((.....)))))...........((((.((........((((.(......).))))))))))))((((((...((((.((((....)))).))))....))))))......... (-32.56 = -31.68 + -0.88)

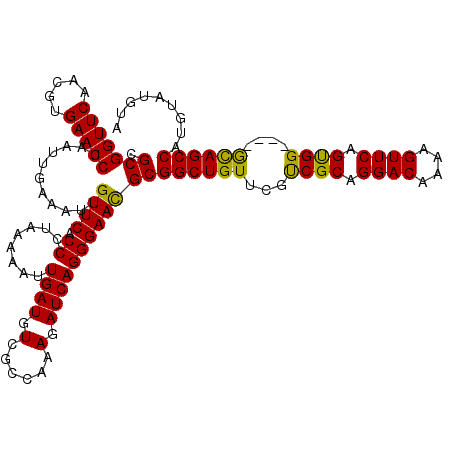

| Location | 8,085,191 – 8,085,311 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -34.24 |
| Consensus MFE | -33.48 |
| Energy contribution | -33.68 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8085191 120 + 27905053 ACGAACAGCCGCAUUCCCUGAUCUUUGGCGACAUCAAUUUUUAGGGUGAACAAUUUCAAUUAGGUUCACGUUGAACCGCGAUUGAGAUUAACGCACUGCAAGUAGUUUCCCGCAGAUAAU ..........((...((((((...((((.....))))...)))))).....((((((((((.(((((.....)))))..))))))))))...)).((((............))))..... ( -31.30) >DroSec_CAF1 10118 120 + 1 ACGAACAGCCGCGUUCCCUGAUCUUUGGCGACAUCAAUUUUUAGGGUGAACAAUUUCAAUUAGGUUCACGUUGAACCGCGAUUGAGAUUAACGCACUGCAAGUAGUUUCCCGCAGAUAAU ..........(((((((((((...((((.....))))...)))))).....((((((((((.(((((.....)))))..))))))))))))))).((((............))))..... ( -34.60) >DroSim_CAF1 8957 120 + 1 ACGAACAGCCGCGUUCCCUGAUCUUUGGCGACAUCAAUUUUUAGGGUGAACAAUUUCAAUUAGGUUCACGUUGAACCGCGAUUGAGAUUAACGCACUGCAAGUAGUUUCCCGCAGAUAAU ..........(((((((((((...((((.....))))...)))))).....((((((((((.(((((.....)))))..))))))))))))))).((((............))))..... ( -34.60) >DroEre_CAF1 11880 119 + 1 ACGAACAGCCGCGUUCCCUGAUCUUUGGCGACAUCAAUUUUUAGGGUGAACAAUUUCAAUUAGGUUCACGUUGAACCGCGAUUGAGAUUAACGCACUGCA-GUAGUUUCCCGCAGAUAAU ..........(((((((((((...((((.....))))...)))))).....((((((((((.(((((.....)))))..))))))))))))))).((((.-..........))))..... ( -34.70) >DroYak_CAF1 5234 120 + 1 GCGAACAGCCGCGUUCCCUGAUCUUUGGCGACAUCAAUUUUUAGGGUGAACAAUUUCAAUUAGGUUCACGUUGAACCGCGAUUGAGAUUAACGCACUGCAAGUAGUUUCCCGCAGAUAAU (((.......(((((((((((...((((.....))))...)))))).....((((((((((.(((((.....)))))..)))))))))))))))((((....))))....)))....... ( -36.00) >consensus ACGAACAGCCGCGUUCCCUGAUCUUUGGCGACAUCAAUUUUUAGGGUGAACAAUUUCAAUUAGGUUCACGUUGAACCGCGAUUGAGAUUAACGCACUGCAAGUAGUUUCCCGCAGAUAAU ..........(((((((((((...((((.....))))...)))))).....((((((((((.(((((.....)))))..))))))))))))))).((((............))))..... (-33.48 = -33.68 + 0.20)

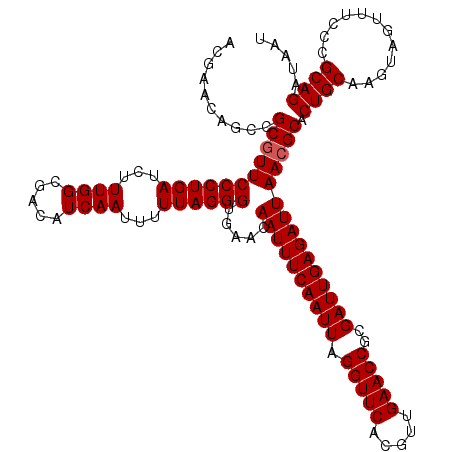
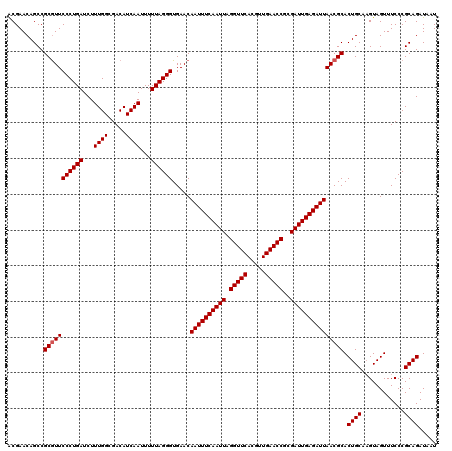
| Location | 8,085,231 – 8,085,351 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.33 |
| Mean single sequence MFE | -36.08 |
| Consensus MFE | -34.62 |
| Energy contribution | -34.82 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8085231 120 + 27905053 UUAGGGUGAACAAUUUCAAUUAGGUUCACGUUGAACCGCGAUUGAGAUUAACGCACUGCAAGUAGUUUCCCGCAGAUAAUGCAAUUGUUGUGUGUAUUUGCGUAUUCCGCACCGCCGGCU ....((((...((((((((((.(((((.....)))))..)))))))))).((((((.(((((((...((.....))...)))..)))).))))))...((((.....)))).)))).... ( -36.30) >DroSec_CAF1 10158 120 + 1 UUAGGGUGAACAAUUUCAAUUAGGUUCACGUUGAACCGCGAUUGAGAUUAACGCACUGCAAGUAGUUUCCCGCAGAUAAUGCAAUUGUUGUGUGUAUUUGCGUAUUCCGCACCGCCGGCU ....((((...((((((((((.(((((.....)))))..)))))))))).((((((.(((((((...((.....))...)))..)))).))))))...((((.....)))).)))).... ( -36.30) >DroSim_CAF1 8997 120 + 1 UUAGGGUGAACAAUUUCAAUUAGGUUCACGUUGAACCGCGAUUGAGAUUAACGCACUGCAAGUAGUUUCCCGCAGAUAAUGCAAUUGUUGUGUGUAUUUGCGUAUUCCGCACCGCCGGCU ....((((...((((((((((.(((((.....)))))..)))))))))).((((((.(((((((...((.....))...)))..)))).))))))...((((.....)))).)))).... ( -36.30) >DroEre_CAF1 11920 119 + 1 UUAGGGUGAACAAUUUCAAUUAGGUUCACGUUGAACCGCGAUUGAGAUUAACGCACUGCA-GUAGUUUCCCGCAGAUAAUGCAAUUGUUGUGUGUAUUUGCGUAUUCCGCACCGCCGGCU ....((((...((((((((((.(((((.....)))))..)))))))))).((((((.(((-((.(.....)(((.....))).))))).))))))...((((.....)))).)))).... ( -37.20) >DroYak_CAF1 5274 120 + 1 UUAGGGUGAACAAUUUCAAUUAGGUUCACGUUGAACCGCGAUUGAGAUUAACGCACUGCAAGUAGUUUCCCGCAGAUAAUGCAAUUGUUGUGUGUAUUUGCGUAUUCCACACCGCCGGCU ....((((...((((((((((.(((((.....)))))..)))))))))).((((((((....)))).....(((.(((((......))))).)))....))))......))))....... ( -34.30) >consensus UUAGGGUGAACAAUUUCAAUUAGGUUCACGUUGAACCGCGAUUGAGAUUAACGCACUGCAAGUAGUUUCCCGCAGAUAAUGCAAUUGUUGUGUGUAUUUGCGUAUUCCGCACCGCCGGCU ....((((...((((((((((.(((((.....)))))..))))))))))...((((((....))))....(((((((((((((.....))))).))))))))......))..)))).... (-34.62 = -34.82 + 0.20)


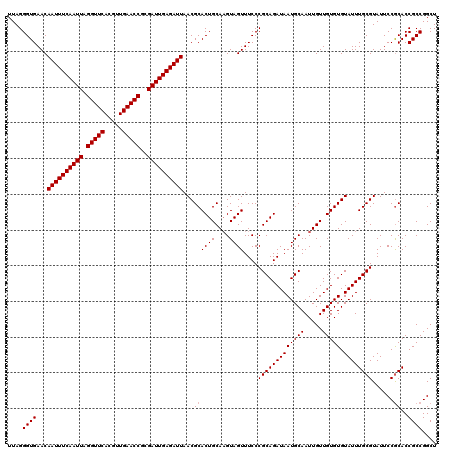
| Location | 8,085,231 – 8,085,351 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.33 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -30.06 |
| Energy contribution | -30.10 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8085231 120 - 27905053 AGCCGGCGGUGCGGAAUACGCAAAUACACACAACAAUUGCAUUAUCUGCGGGAAACUACUUGCAGUGCGUUAAUCUCAAUCGCGGUUCAACGUGAACCUAAUUGAAAUUGUUCACCCUAA ....((.((((((.....)))..........((((((((((....(((((((......)))))))))).......(((((...(((((.....)))))..)))))))))))).))))).. ( -31.00) >DroSec_CAF1 10158 120 - 1 AGCCGGCGGUGCGGAAUACGCAAAUACACACAACAAUUGCAUUAUCUGCGGGAAACUACUUGCAGUGCGUUAAUCUCAAUCGCGGUUCAACGUGAACCUAAUUGAAAUUGUUCACCCUAA ....((.((((((.....)))..........((((((((((....(((((((......)))))))))).......(((((...(((((.....)))))..)))))))))))).))))).. ( -31.00) >DroSim_CAF1 8997 120 - 1 AGCCGGCGGUGCGGAAUACGCAAAUACACACAACAAUUGCAUUAUCUGCGGGAAACUACUUGCAGUGCGUUAAUCUCAAUCGCGGUUCAACGUGAACCUAAUUGAAAUUGUUCACCCUAA ....((.((((((.....)))..........((((((((((....(((((((......)))))))))).......(((((...(((((.....)))))..)))))))))))).))))).. ( -31.00) >DroEre_CAF1 11920 119 - 1 AGCCGGCGGUGCGGAAUACGCAAAUACACACAACAAUUGCAUUAUCUGCGGGAAACUAC-UGCAGUGCGUUAAUCUCAAUCGCGGUUCAACGUGAACCUAAUUGAAAUUGUUCACCCUAA ....((.((((((.....)))..........((((((((((....((((((.......)-)))))))).......(((((...(((((.....)))))..)))))))))))).))))).. ( -31.30) >DroYak_CAF1 5274 120 - 1 AGCCGGCGGUGUGGAAUACGCAAAUACACACAACAAUUGCAUUAUCUGCGGGAAACUACUUGCAGUGCGUUAAUCUCAAUCGCGGUUCAACGUGAACCUAAUUGAAAUUGUUCACCCUAA ....((.((((((.............)))))((((((((((....(((((((......)))))))))).......(((((...(((((.....)))))..))))))))))))).)).... ( -30.72) >consensus AGCCGGCGGUGCGGAAUACGCAAAUACACACAACAAUUGCAUUAUCUGCGGGAAACUACUUGCAGUGCGUUAAUCUCAAUCGCGGUUCAACGUGAACCUAAUUGAAAUUGUUCACCCUAA ....((.((((((.....)))..........((((((((((....(((((((......)))))))))).......(((((...(((((.....)))))..)))))))))))).))))).. (-30.06 = -30.10 + 0.04)

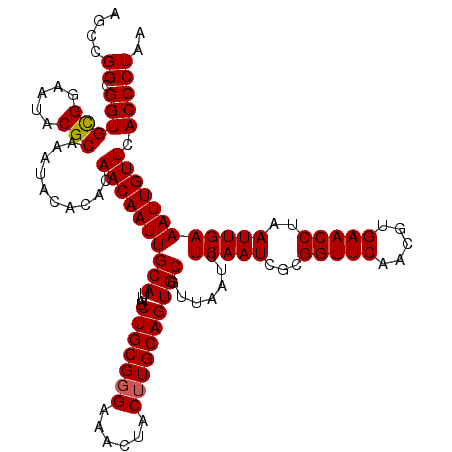
| Location | 8,085,311 – 8,085,431 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -38.80 |
| Consensus MFE | -38.42 |
| Energy contribution | -38.46 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8085311 120 - 27905053 CAUCUAUUGUCCCUCGCCAAUCGACGGCCAUUGUCUUUCGAUUCUAGGCGUGGGAUACUGCUAACCGUUUGGCGUUUUCCAGCCGGCGGUGCGGAAUACGCAAAUACACACAACAAUUGC .......((((((.((((((((((.(((....)))..))))))...)))).))))))((((..((((((.(((........))))))))))))).....((((.............)))) ( -39.92) >DroSec_CAF1 10238 120 - 1 CAUCUAUUGUCCCUCGCCAAUCGACGGCCUUUGUCUUUCGAUUCGAGGCGUGGGAUACUGCUAACCGUUUGGCGUUUUCCAGCCGGCGGUGCGGAAUACGCAAAUACACACAACAAUUGC .......((((((.((((((((((.(((....)))..))))))...)))).))))))((((..((((((.(((........))))))))))))).....((((.............)))) ( -39.92) >DroSim_CAF1 9077 120 - 1 CAUCUAUUGUCCCUCGCCAAUCGACGGCCUUUGUCUUUCGAUUCGAGGCGUGGGAUACUGCUAACCGUUUGGCGUUUUCCAGCCGGCGGUGCGGAAUACGCAAAUACACACAACAAUUGC .......((((((.((((((((((.(((....)))..))))))...)))).))))))((((..((((((.(((........))))))))))))).....((((.............)))) ( -39.92) >DroEre_CAF1 11999 119 - 1 CAUCUAUUGUCCCUCGCCAAUCGACGGCCUUUGUCUUUCGAUUCGAGGCGUGGGAUACUGCUAACCGUUUGGCGUUUU-CAGCCGGCGGUGCGGAAUACGCAAAUACACACAACAAUUGC .......((((((.((((((((((.(((....)))..))))))...)))).))))))((((..((((((.(((.....-..))))))))))))).....((((.............)))) ( -39.62) >DroYak_CAF1 5354 120 - 1 CAUCUAUUGUCCCUCGCCAAUCGACGCCCAUUGUCUUUCGAUUCGAGGCGUGGGAUACUGCUAACCGUUUGGCAUUUUCCAGCCGGCGGUGUGGAAUACGCAAAUACACACAACAAUUGC ..(((((((((((.((((((((((.((.....))...))))))...)))).))))))......((((((.(((........))))))))))))))....((((.............)))) ( -34.62) >consensus CAUCUAUUGUCCCUCGCCAAUCGACGGCCUUUGUCUUUCGAUUCGAGGCGUGGGAUACUGCUAACCGUUUGGCGUUUUCCAGCCGGCGGUGCGGAAUACGCAAAUACACACAACAAUUGC .......((((((.((((((((((.(((....)))..))))))...)))).))))))((((..((((((.(((........))))))))))))).....((((.............)))) (-38.42 = -38.46 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:54 2006