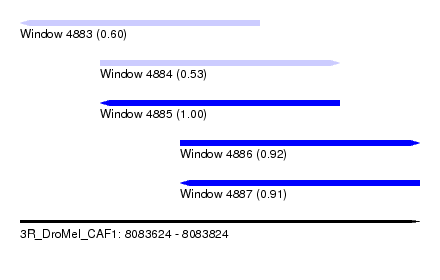
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,083,624 – 8,083,824 |
| Length | 200 |
| Max. P | 0.995005 |

| Location | 8,083,624 – 8,083,744 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -45.90 |
| Consensus MFE | -43.82 |
| Energy contribution | -43.58 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8083624 120 - 27905053 ACCAAGGGAAUGCAAUGGGCUGGAAGCGACCGGGAUGGGGUGGUCAUCCGAUCGCCCCGAAGACAGCUGCCGUGUGCUGGAACCGGACAAUGUCCAAGGGACCCAUGGGCCACAAAAGUU .(((.(((..(.(..(((((((....)).((((..((((((((((....))))))))))....((((........))))...)))).....)))))..).)))).)))............ ( -45.10) >DroSec_CAF1 8567 120 - 1 ACCAAGGGAAUGCAAUGGGCUGGAAGCGACCGGGAUGGGGUGGUCAUCCGAUCGCCCCGAAGACAGCUGCCGUGUGCUGGAACCGGACAAUAUCCAAGGGGCCCAUGGGCCACAAAAGUU .((...(((.........((.....))..((((..((((((((((....))))))))))....((((........))))...))))......)))..))((((....))))......... ( -46.40) >DroSim_CAF1 6984 120 - 1 ACCAAGGGAAUGCAAUGGGCUGGAAGCGACCGGGAUGGGGUGGUCAUCCGAUCGCCCCGAAGACAGCUGCCGUGUGCUGGAACCGGACAACGUCCAAGGGACCCAUGGGCCACAAAAGUC .(((.(((..(.(..(((((((....)).((((..((((((((((....))))))))))....((((........))))...)))).....)))))..).)))).)))............ ( -45.40) >DroEre_CAF1 10343 120 - 1 ACCAAGGGAAUGCAAUGGGCUGGAAGCGACCGGGAUGGGGUGGUCAUCCGAUCGCCCCGAAGACAGCUGCCGUGUGCUGGAACCGGCCAAUCUCCGAGGGACCCAUGGGCCACAAAAGUU .(((.(((.........((((((.((((..(((..((((((((((....)))))))))).((....)).)))..))))....))))))..(((.....)))))).)))............ ( -47.00) >DroYak_CAF1 3684 119 - 1 ACCAAGGGAAUGCAAUGGGCUGGAAGCGACCGGGGUGGGGUGGUCAUCCGAUCGCCCCGAAGACAGCUGCCGUGUGCUGGAACCGGACCAUAUCCGAG-GACCCAUGGGCCACAAAAGUU .(((.(((......((((((.....))..((((..((((((((((....))))))))))....((((........))))...)))).)))).((....-))))).)))............ ( -45.60) >consensus ACCAAGGGAAUGCAAUGGGCUGGAAGCGACCGGGAUGGGGUGGUCAUCCGAUCGCCCCGAAGACAGCUGCCGUGUGCUGGAACCGGACAAUAUCCAAGGGACCCAUGGGCCACAAAAGUU .(((.(((...((.....))((((.....((((..((((((((((....))))))))))....((((........))))...))))......)))).....))).)))............ (-43.82 = -43.58 + -0.24)


| Location | 8,083,664 – 8,083,784 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.67 |
| Mean single sequence MFE | -38.46 |
| Consensus MFE | -38.00 |
| Energy contribution | -38.20 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.99 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8083664 120 + 27905053 CCAGCACACGGCAGCUGUCUUCGGGGCGAUCGGAUGACCACCCCAUCCCGGUCGCUUCCAGCCCAUUGCAUUCCCUUGGUCUGGUCUUUGUCCGGGAUUGUUUGCCUUCAAUUUGACUAC .........((((((((.....((((((((((((((.......))).))))))))))))))).....(((.((((..((.(........).)))))).))).)))).............. ( -38.90) >DroSec_CAF1 8607 120 + 1 CCAGCACACGGCAGCUGUCUUCGGGGCGAUCGGAUGACCACCCCAUCCCGGUCGCUUCCAGCCCAUUGCAUUCCCUUGGUCUGGUCUUUGUCCGGGAUUGUUUGCCUUCAAUUUGACUAC .........((((((((.....((((((((((((((.......))).))))))))))))))).....(((.((((..((.(........).)))))).))).)))).............. ( -38.90) >DroSim_CAF1 7024 120 + 1 CCAGCACACGGCAGCUGUCUUCGGGGCGAUCGGAUGACCACCCCAUCCCGGUCGCUUCCAGCCCAUUGCAUUCCCUUGGUCUGGUCUUUGUCCGGGAUUGUUUGCCUUCAAUUUGACUAC .........((((((((.....((((((((((((((.......))).))))))))))))))).....(((.((((..((.(........).)))))).))).)))).............. ( -38.90) >DroEre_CAF1 10383 120 + 1 CCAGCACACGGCAGCUGUCUUCGGGGCGAUCGGAUGACCACCCCAUCCCGGUCGCUUCCAGCCCAUUGCAUUCCCUUGGUCUGGUCUUUGUCCGGGAUUGUUUGCCUUCAAUUUGACUAC .........((((((((.....((((((((((((((.......))).))))))))))))))).....(((.((((..((.(........).)))))).))).)))).............. ( -38.90) >DroYak_CAF1 3723 120 + 1 CCAGCACACGGCAGCUGUCUUCGGGGCGAUCGGAUGACCACCCCACCCCGGUCGCUUCCAGCCCAUUGCAUUCCCUUGGUCUGGUCUUUGUCCGGGAUUGUUUGCCUUCAAUUUGACUAC .........((((((((.....(((((((((((.((.......))..))))))))))))))).....(((.((((..((.(........).)))))).))).)))).............. ( -36.70) >consensus CCAGCACACGGCAGCUGUCUUCGGGGCGAUCGGAUGACCACCCCAUCCCGGUCGCUUCCAGCCCAUUGCAUUCCCUUGGUCUGGUCUUUGUCCGGGAUUGUUUGCCUUCAAUUUGACUAC .........((((((((.....((((((((((((((.......))).))))))))))))))).....(((.((((..((.(........).)))))).))).)))).............. (-38.00 = -38.20 + 0.20)

| Location | 8,083,664 – 8,083,784 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.67 |
| Mean single sequence MFE | -45.00 |
| Consensus MFE | -45.16 |
| Energy contribution | -45.00 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.05 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.995005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8083664 120 - 27905053 GUAGUCAAAUUGAAGGCAAACAAUCCCGGACAAAGACCAGACCAAGGGAAUGCAAUGGGCUGGAAGCGACCGGGAUGGGGUGGUCAUCCGAUCGCCCCGAAGACAGCUGCCGUGUGCUGG ((..((.....))..)).....(((((((.......((((.(((..(.....)..))).))))......)))))))(((((((((....))))))))).....((((........)))). ( -45.12) >DroSec_CAF1 8607 120 - 1 GUAGUCAAAUUGAAGGCAAACAAUCCCGGACAAAGACCAGACCAAGGGAAUGCAAUGGGCUGGAAGCGACCGGGAUGGGGUGGUCAUCCGAUCGCCCCGAAGACAGCUGCCGUGUGCUGG ((..((.....))..)).....(((((((.......((((.(((..(.....)..))).))))......)))))))(((((((((....))))))))).....((((........)))). ( -45.12) >DroSim_CAF1 7024 120 - 1 GUAGUCAAAUUGAAGGCAAACAAUCCCGGACAAAGACCAGACCAAGGGAAUGCAAUGGGCUGGAAGCGACCGGGAUGGGGUGGUCAUCCGAUCGCCCCGAAGACAGCUGCCGUGUGCUGG ((..((.....))..)).....(((((((.......((((.(((..(.....)..))).))))......)))))))(((((((((....))))))))).....((((........)))). ( -45.12) >DroEre_CAF1 10383 120 - 1 GUAGUCAAAUUGAAGGCAAACAAUCCCGGACAAAGACCAGACCAAGGGAAUGCAAUGGGCUGGAAGCGACCGGGAUGGGGUGGUCAUCCGAUCGCCCCGAAGACAGCUGCCGUGUGCUGG ((..((.....))..)).....(((((((.......((((.(((..(.....)..))).))))......)))))))(((((((((....))))))))).....((((........)))). ( -45.12) >DroYak_CAF1 3723 120 - 1 GUAGUCAAAUUGAAGGCAAACAAUCCCGGACAAAGACCAGACCAAGGGAAUGCAAUGGGCUGGAAGCGACCGGGGUGGGGUGGUCAUCCGAUCGCCCCGAAGACAGCUGCCGUGUGCUGG ((..((.....))..)).....(((((((.......((((.(((..(.....)..))).))))......)))))))(((((((((....))))))))).....((((........)))). ( -44.52) >consensus GUAGUCAAAUUGAAGGCAAACAAUCCCGGACAAAGACCAGACCAAGGGAAUGCAAUGGGCUGGAAGCGACCGGGAUGGGGUGGUCAUCCGAUCGCCCCGAAGACAGCUGCCGUGUGCUGG ((..((.....))..)).....(((((((.......((((.(((..(.....)..))).))))......)))))))(((((((((....))))))))).....((((........)))). (-45.16 = -45.00 + -0.16)

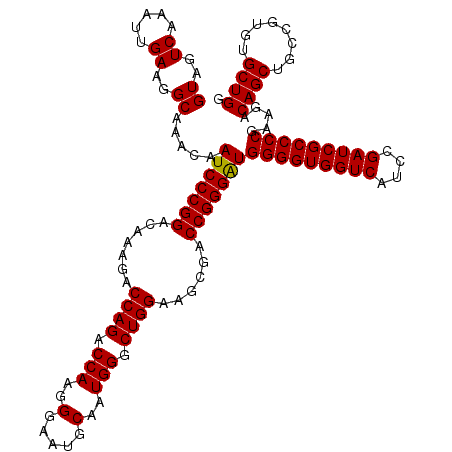
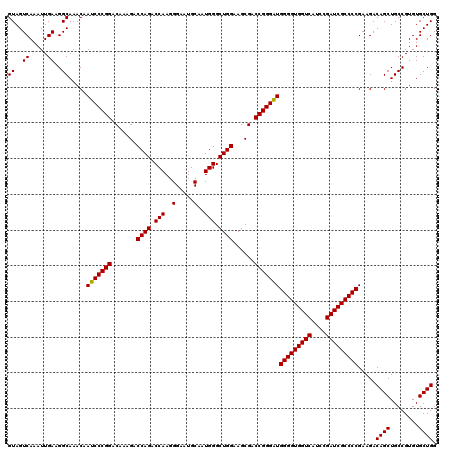
| Location | 8,083,704 – 8,083,824 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.67 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -30.60 |
| Energy contribution | -30.80 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8083704 120 + 27905053 CCCCAUCCCGGUCGCUUCCAGCCCAUUGCAUUCCCUUGGUCUGGUCUUUGUCCGGGAUUGUUUGCCUUCAAUUUGACUACAAACCAUAAUUUAUCUUUUCAGACGUGUUGGUCAGCGCGA ....(((((((.((...((((.(((...........))).))))....)).))))))).(((((...((.....))...)))))...................((((((....)))))). ( -31.50) >DroSec_CAF1 8647 120 + 1 CCCCAUCCCGGUCGCUUCCAGCCCAUUGCAUUCCCUUGGUCUGGUCUUUGUCCGGGAUUGUUUGCCUUCAAUUUGACUACAAACCAUAAUUUAUCUUUUCAGACGUGUUGGUCAGCGCGA ....(((((((.((...((((.(((...........))).))))....)).))))))).(((((...((.....))...)))))...................((((((....)))))). ( -31.50) >DroSim_CAF1 7064 120 + 1 CCCCAUCCCGGUCGCUUCCAGCCCAUUGCAUUCCCUUGGUCUGGUCUUUGUCCGGGAUUGUUUGCCUUCAAUUUGACUACAAACCAUAAUUUAUCUUUUCAGACGUGUUGGUCAGCGCGA ....(((((((.((...((((.(((...........))).))))....)).))))))).(((((...((.....))...)))))...................((((((....)))))). ( -31.50) >DroEre_CAF1 10423 120 + 1 CCCCAUCCCGGUCGCUUCCAGCCCAUUGCAUUCCCUUGGUCUGGUCUUUGUCCGGGAUUGUUUGCCUUCAAUUUGACUACAAACCAUAAUUUAUCUUUUCAGACGUGUUGGUCAGCGCGA ....(((((((.((...((((.(((...........))).))))....)).))))))).(((((...((.....))...)))))...................((((((....)))))). ( -31.50) >DroYak_CAF1 3763 120 + 1 CCCCACCCCGGUCGCUUCCAGCCCAUUGCAUUCCCUUGGUCUGGUCUUUGUCCGGGAUUGUUUGCCUUCAAUUUGACUACAAACCAUAAUUUAUCUUUUCAGACGUGUUGGUCAGCGCGA ......(((((.((...((((.(((...........))).))))....)).)))))...(((((...((.....))...)))))...................((((((....)))))). ( -29.50) >consensus CCCCAUCCCGGUCGCUUCCAGCCCAUUGCAUUCCCUUGGUCUGGUCUUUGUCCGGGAUUGUUUGCCUUCAAUUUGACUACAAACCAUAAUUUAUCUUUUCAGACGUGUUGGUCAGCGCGA ....(((((((.((...((((.(((...........))).))))....)).))))))).(((((...((.....))...)))))...................((((((....)))))). (-30.60 = -30.80 + 0.20)

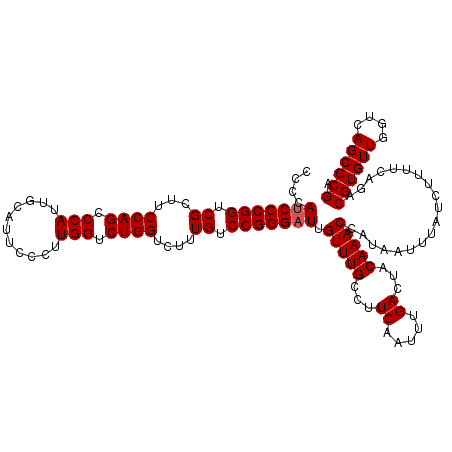
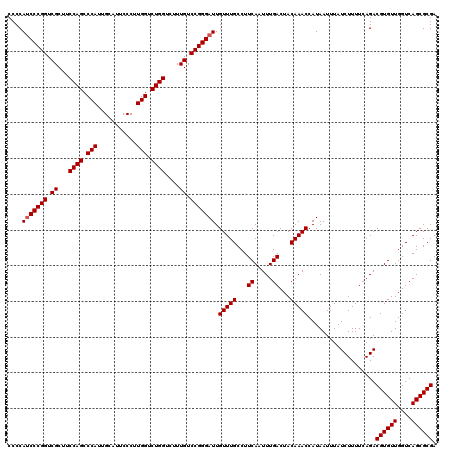
| Location | 8,083,704 – 8,083,824 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.67 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -34.26 |
| Energy contribution | -34.10 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.53 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8083704 120 - 27905053 UCGCGCUGACCAACACGUCUGAAAAGAUAAAUUAUGGUUUGUAGUCAAAUUGAAGGCAAACAAUCCCGGACAAAGACCAGACCAAGGGAAUGCAAUGGGCUGGAAGCGACCGGGAUGGGG .........((.....((((....))))........((((((..((.....))..)))))).(((((((.......((((.(((..(.....)..))).))))......))))))))).. ( -34.22) >DroSec_CAF1 8647 120 - 1 UCGCGCUGACCAACACGUCUGAAAAGAUAAAUUAUGGUUUGUAGUCAAAUUGAAGGCAAACAAUCCCGGACAAAGACCAGACCAAGGGAAUGCAAUGGGCUGGAAGCGACCGGGAUGGGG .........((.....((((....))))........((((((..((.....))..)))))).(((((((.......((((.(((..(.....)..))).))))......))))))))).. ( -34.22) >DroSim_CAF1 7064 120 - 1 UCGCGCUGACCAACACGUCUGAAAAGAUAAAUUAUGGUUUGUAGUCAAAUUGAAGGCAAACAAUCCCGGACAAAGACCAGACCAAGGGAAUGCAAUGGGCUGGAAGCGACCGGGAUGGGG .........((.....((((....))))........((((((..((.....))..)))))).(((((((.......((((.(((..(.....)..))).))))......))))))))).. ( -34.22) >DroEre_CAF1 10423 120 - 1 UCGCGCUGACCAACACGUCUGAAAAGAUAAAUUAUGGUUUGUAGUCAAAUUGAAGGCAAACAAUCCCGGACAAAGACCAGACCAAGGGAAUGCAAUGGGCUGGAAGCGACCGGGAUGGGG .........((.....((((....))))........((((((..((.....))..)))))).(((((((.......((((.(((..(.....)..))).))))......))))))))).. ( -34.22) >DroYak_CAF1 3763 120 - 1 UCGCGCUGACCAACACGUCUGAAAAGAUAAAUUAUGGUUUGUAGUCAAAUUGAAGGCAAACAAUCCCGGACAAAGACCAGACCAAGGGAAUGCAAUGGGCUGGAAGCGACCGGGGUGGGG .........((.....((((....))))........((((((..((.....))..)))))).(((((((.......((((.(((..(.....)..))).))))......))))))))).. ( -33.62) >consensus UCGCGCUGACCAACACGUCUGAAAAGAUAAAUUAUGGUUUGUAGUCAAAUUGAAGGCAAACAAUCCCGGACAAAGACCAGACCAAGGGAAUGCAAUGGGCUGGAAGCGACCGGGAUGGGG .........((.....((((....))))........((((((..((.....))..)))))).(((((((.......((((.(((..(.....)..))).))))......))))))))).. (-34.26 = -34.10 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:47 2006