
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,080,136 – 8,080,290 |
| Length | 154 |
| Max. P | 0.870178 |

| Location | 8,080,136 – 8,080,256 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.44 |
| Mean single sequence MFE | -40.82 |
| Consensus MFE | -26.09 |
| Energy contribution | -25.43 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.619154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8080136 120 - 27905053 GUACACCUCCUUGGGAUCCCAGUUUCCAAGAGAGGGACAGUUGCCACUAAUGGCAACAUACUCAUUACCGUUUAGUGAACUGUGAAAGACUACACAGAAGGGCCACUCAGUAGUCGGAAC ((...(((((((((((.......))))))).)))).)).(((((((....)))))))..........(((.(((.(((.(((((........)))))..(.....)))).))).)))... ( -39.40) >DroVir_CAF1 15511 120 - 1 GUACACCUGCUUCGGGUCCCAGUUGCCCAGCUCGUCGCAGUUGCCGCUGAUCGCCAUAUGCUCGUUGCCCUGCAGCGGCAUCUUGAUCCGCACACAAAAUGGCCAAUCCGUAUAGACAAU ((((..((((..(((((............)))))..))))..(((((.(((((....((((.((((((...))))))))))..))))).)).........)))......))))....... ( -32.60) >DroSec_CAF1 5056 120 - 1 GUACACCUCCUUGGGAUCCCAGUUUCCGAGAGAGGGACAGUUGCCACUAAUGGCAACAUACUCAUUUCCGUUGAGUGAACUGUGGAACACCACACAGAAUGGCCACUCGGUAGUCGGAAC ((...(((((((((((.......))))))).)))).)).(((((((....)))))))........((((((((((((..((((((....)))))......)..)))))))....))))). ( -43.40) >DroSim_CAF1 3504 120 - 1 GUACACCUCCUUGGGAUCCCAGUUUCCGAGAGAGGGACAGUUGCCACUAAUGGCAACAUACUCAUUUCCGUUGAGUGAACUGUGGAACACCACACAGAAGGGCCACUCGGUAGUCGGAAC ((...(((((((((((.......))))))).)))).)).(((((((....)))))))........((((((((((((..((((((....))))......))..)))))))....))))). ( -43.40) >DroEre_CAF1 6754 120 - 1 GUACACCUUCUUGGGAUCCCAGUUCCCGAGAGACGGACAGUUGCCACUAAUGGCCACAUACUCAUUCCCUUUGAGUGGAUGGUGGAACACCACACAGAAGGGCCACUCUGUAGUCGGCAC .....(((((((((((.......)))))))))..)).....((((((((.(((((...((((((.......))))))..((((((....)))).))....))))).....)))).)))). ( -42.30) >DroYak_CAF1 109 120 - 1 GAACACCUUCUUGGGAUCCCAGUUCCCGAGAGAGGGACAGUUGCCACUUAUGGCCACAUACUCAUUCCCUUUGAGUGGAUGGUGGUACACCACACAGAAGGGCCACUCUGUGGUCGGAAC .....(((((((((((.......)))))))).))).......(((((...(((((...((((((.......))))))..((((((....)))).))....)))))....)))))...... ( -43.80) >consensus GUACACCUCCUUGGGAUCCCAGUUUCCGAGAGAGGGACAGUUGCCACUAAUGGCAACAUACUCAUUCCCGUUGAGUGAACUGUGGAACACCACACAGAAGGGCCACUCGGUAGUCGGAAC .....(((((((((((.......))))))).))))(((.((.((((....)))).)).(((((((((.....)))))).(((((........)))))............))))))..... (-26.09 = -25.43 + -0.66)



| Location | 8,080,176 – 8,080,290 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.25 |
| Mean single sequence MFE | -37.68 |
| Consensus MFE | -23.35 |
| Energy contribution | -23.08 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8080176 114 - 27905053 AUA------GGCAGCUAAUGCAGUCAUUCUUGGCCAAAAUGUACACCUCCUUGGGAUCCCAGUUUCCAAGAGAGGGACAGUUGCCACUAAUGGCAACAUACUCAUUACCGUUUAGUGAAC ...------(((.((....))...((....)))))....(((...(((((((((((.......))))))).)))).)))(((((((....)))))))....((((((.....)))))).. ( -37.00) >DroVir_CAF1 15551 117 - 1 ACUGGCCCGCACAGG---UGCAGUUCGCCGGAUCCAUAAUGUACACCUGCUUCGGGUCCCAGUUGCCCAGCUCGUCGCAGUUGCCGCUGAUCGCCAUAUGCUCGUUGCCCUGCAGCGGCA (((((((((..((((---((..(((....(....)..)))...))))))...))))..)))))......((.....))...((((((((...((.((......)).))....)))))))) ( -35.60) >DroSec_CAF1 5096 114 - 1 AUA------GGCAGUUCAUGCAGUCAUGCUUUGCCAAAAUGUACACCUCCUUGGGAUCCCAGUUUCCGAGAGAGGGACAGUUGCCACUAAUGGCAACAUACUCAUUUCCGUUGAGUGAAC ...------(((((..((((....))))..)))))....(((...(((((((((((.......))))))).)))).)))(((((((....))))))).((((((.......))))))... ( -43.80) >DroSim_CAF1 3544 114 - 1 AUA------GGCAGUUCAUGCAGUCAUGCUUGGCCAAAAUGUACACCUCCUUGGGAUCCCAGUUUCCGAGAGAGGGACAGUUGCCACUAAUGGCAACAUACUCAUUUCCGUUGAGUGAAC ...------(((....((((....))))....)))....(((...(((((((((((.......))))))).)))).)))(((((((....))))))).((((((.......))))))... ( -40.70) >DroEre_CAF1 6794 114 - 1 AUA------GGCAGUUGAUGCAGUCGUUCCUUUCCAAGAUGUACACCUUCUUGGGAUCCCAGUUCCCGAGAGACGGACAGUUGCCACUAAUGGCCACAUACUCAUUCCCUUUGAGUGGAU ...------(((.((((..((((..((((((.....)).........(((((((((.......)))))))))..))))..))))...)))).)))...((((((.......))))))... ( -31.60) >DroYak_CAF1 149 114 - 1 AUA------GGCAGUUGAUGCACUCGUGCUUUGCCAGGAUGAACACCUUCUUGGGAUCCCAGUUCCCGAGAGAGGGACAGUUGCCACUUAUGGCCACAUACUCAUUCCCUUUGAGUGGAU ...------(((((.....(((....))).))))).((((((...(((((((((((.......)))))))).)))....((.((((....)))).))....))))))((.......)).. ( -37.40) >consensus AUA______GGCAGUUCAUGCAGUCAUGCUUUGCCAAAAUGUACACCUCCUUGGGAUCCCAGUUUCCGAGAGAGGGACAGUUGCCACUAAUGGCAACAUACUCAUUCCCGUUGAGUGAAC .........(((....((((....))))....)))....(((...(((((((((((.......))))))).)))).)))((.((((....)))).))....((((((.....)))))).. (-23.35 = -23.08 + -0.27)

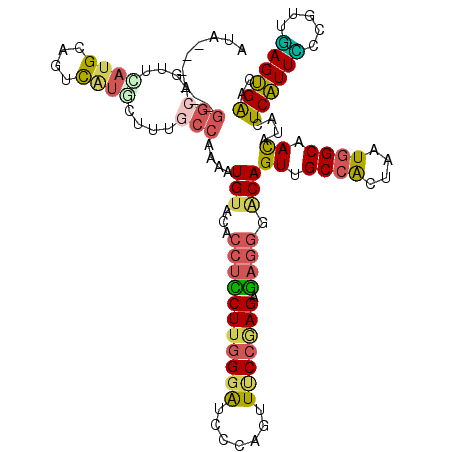

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:37 2006