
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,075,879 – 8,075,996 |
| Length | 117 |
| Max. P | 0.700615 |

| Location | 8,075,879 – 8,075,996 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -26.97 |
| Consensus MFE | -17.07 |
| Energy contribution | -18.07 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
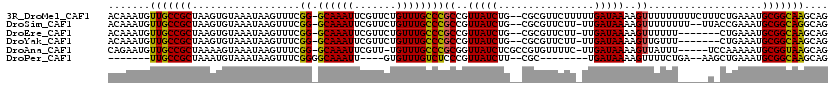
>3R_DroMel_CAF1 8075879 117 + 27905053 CUGCUUGCCGCAUUUCAGAAAGAAAAAAAAACUUUUAUCAAAAAGAACGCG--CAGAUAACGGCGGGCAAACAGAACGAAUUUGC-CCGAAACUUAUUUACACUUAGCGGCAACAUUUGU .((.(((((((.((((.....))))......(((((....))))).....(--.((((((.(.((((((((.........)))))-)))...))))))).).....)))))))))..... ( -29.00) >DroSim_CAF1 21928 114 + 1 CUGCCUGCCGCAUUUCGGUAA--AAAAAAAACUUUUAUCAA-AAGAACGCG--CAGAUAACGGCGGGCAAACAGAACGAAUUUGC-CCGAAACUUAUUUACACUUAGCGGCAACAUUUGU .....((((((.....(((((--((.......)))))))..-........(--.((((((.(.((((((((.........)))))-)))...))))))).).....))))))........ ( -29.70) >DroEre_CAF1 20438 109 + 1 CUGCUUGCCGCAUUUCAG-------AAAAAACUUUUAUCAA-AAGAACGCG--CAGAUAACGGCGGGCAAACAGAACGAAUUUGC-CCGAAACUUAUUUACACUUAGCGGCAACAUUUGU .((.(((((((.......-------......((((.....)-))).....(--.((((((.(.((((((((.........)))))-)))...))))))).).....)))))))))..... ( -27.60) >DroYak_CAF1 20855 109 + 1 CUGCUUGCCGCAUUUCAG-------AAACAACUUUUAUCAA-AAGAACGCG--CAGAUAACGGCGGGCAAACAGAACGAAUUUGC-CCGAAACUUAUUUACACUUAGCGGCAACAUUUGU .((.(((((((.......-------......((((.....)-))).....(--.((((((.(.((((((((.........)))))-)))...))))))).).....)))))))))..... ( -27.60) >DroAna_CAF1 27298 112 + 1 CUGCUUACCGCAUUUUUGGA-----AAAUAACUUUUAUCAA-GAAAACACGGCGAGAUAACCGCGGGCAAACA-AACGAAUUUGC-CCGAAACUUAUUUACUUUUAGCGGCAACAUUCUG .......((((.(((((((.-----(((.....))).))))-))).........((((((...((((((((..-......)))))-)))....)))))).......)))).......... ( -25.10) >DroPer_CAF1 22068 97 + 1 CUGCUUGCCGCAUUUCAGCUU--UCAGAAAACUUUUAUCA--------GCG--AAGAUAACGGGAGACAAACAC----AAUUUGCCCCGAAACUUAUUUACAUUUAGCGGCAA------- .....((((((.((((.....--...))))....(((((.--------...--..)))))((((.(.((((...----..))))))))).................)))))).------- ( -22.80) >consensus CUGCUUGCCGCAUUUCAG_A_____AAAAAACUUUUAUCAA_AAGAACGCG__CAGAUAACGGCGGGCAAACAGAACGAAUUUGC_CCGAAACUUAUUUACACUUAGCGGCAACAUUUGU ....(((((((....................((.(((((................))))).))((((((((.........))))).))).................)))))))....... (-17.07 = -18.07 + 1.00)



| Location | 8,075,879 – 8,075,996 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -22.03 |
| Energy contribution | -21.64 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
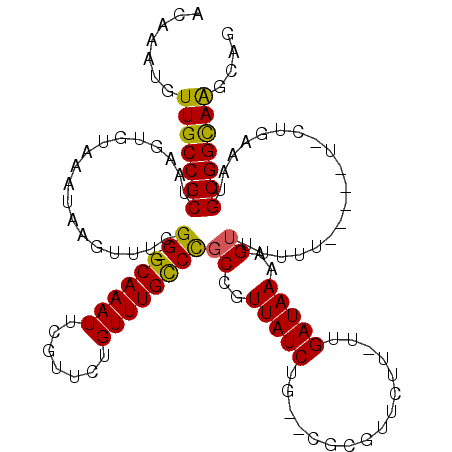
>3R_DroMel_CAF1 8075879 117 - 27905053 ACAAAUGUUGCCGCUAAGUGUAAAUAAGUUUCGG-GCAAAUUCGUUCUGUUUGCCCGCCGUUAUCUG--CGCGUUCUUUUUGAUAAAAGUUUUUUUUUCUUUCUGAAAUGCGGCAAGCAG .....(((((((((...(((((.((((....(((-((((((.......)))))))))...)))).))--)))...(((((....)))))......((((.....)))).))))).)))). ( -34.50) >DroSim_CAF1 21928 114 - 1 ACAAAUGUUGCCGCUAAGUGUAAAUAAGUUUCGG-GCAAAUUCGUUCUGUUUGCCCGCCGUUAUCUG--CGCGUUCUU-UUGAUAAAAGUUUUUUUU--UUACCGAAAUGCGGCAGGCAG .....(((((((((...(((((.((((....(((-((((((.......)))))))))...)))).))--)))....((-(((.((((((.....)))--))).))))).)))))).))). ( -37.00) >DroEre_CAF1 20438 109 - 1 ACAAAUGUUGCCGCUAAGUGUAAAUAAGUUUCGG-GCAAAUUCGUUCUGUUUGCCCGCCGUUAUCUG--CGCGUUCUU-UUGAUAAAAGUUUUUU-------CUGAAAUGCGGCAAGCAG .....(((((((((...(((((.((((....(((-((((((.......)))))))))...)))).))--)))......-.........(((((..-------..)))))))))).)))). ( -33.50) >DroYak_CAF1 20855 109 - 1 ACAAAUGUUGCCGCUAAGUGUAAAUAAGUUUCGG-GCAAAUUCGUUCUGUUUGCCCGCCGUUAUCUG--CGCGUUCUU-UUGAUAAAAGUUGUUU-------CUGAAAUGCGGCAAGCAG .....(((((((((...(((((.((((....(((-((((((.......)))))))))...)))).))--))).(((..-..((((.....)))).-------..)))..))))).)))). ( -33.50) >DroAna_CAF1 27298 112 - 1 CAGAAUGUUGCCGCUAAAAGUAAAUAAGUUUCGG-GCAAAUUCGUU-UGUUUGCCCGCGGUUAUCUCGCCGUGUUUUC-UUGAUAAAAGUUAUUU-----UCCAAAAAUGCGGUAAGCAG .....(((((((((.......((((((.(((.((-((((((.....-.))))))))(((((......)))))......-......))).))))))-----.........))))).)))). ( -32.49) >DroPer_CAF1 22068 97 - 1 -------UUGCCGCUAAAUGUAAAUAAGUUUCGGGGCAAAUU----GUGUUUGUCUCCCGUUAUCUU--CGC--------UGAUAAAAGUUUUCUGA--AAGCUGAAAUGCGGCAAGCAG -------(((((((...((....))..((((((((((((((.----..)))))))))...(((((..--...--------.))))).(((((.....--))))))))))))))))).... ( -24.50) >consensus ACAAAUGUUGCCGCUAAGUGUAAAUAAGUUUCGG_GCAAAUUCGUUCUGUUUGCCCGCCGUUAUCUG__CGCGUUCUU_UUGAUAAAAGUUUUUU_____U_CUGAAAUGCGGCAAGCAG .......(((((((..................((.((((((.......))))))))((..(((((................)))))..))...................))))))).... (-22.03 = -21.64 + -0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:28 2006