
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,073,228 – 8,073,388 |
| Length | 160 |
| Max. P | 0.962661 |

| Location | 8,073,228 – 8,073,319 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.54 |
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -21.16 |
| Energy contribution | -20.97 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8073228 91 + 27905053 UUGUGUUUGACAAAGUUGCGAAACAAGAUGUUAACCCCGGGGGUUGACUCCA---------------GAAUGAAGGCAGGGAUGCAGCAGCAUCUUGAGAAAAGAU-------------- ((((.....)))).(((((.......((.((((((((...))))))))))..---------------........(((....))).)))))(((((.....)))))-------------- ( -23.40) >DroPse_CAF1 19166 119 + 1 UUGUGUUUGACAAAGUUGCGCAACAAGAUGUUAACCCCGAGGGUUGACAUCUGUACUCCGUACAACGUGGUGGA-GUGGGGACGCAACAGCAUCUUGAGAAAAGAUCUCAGAGAUCUCUG ((((.....)))).(((((((..(.((((((((((((...))))))))))))..((((((..........))))-)))..).))))))((.(((((((((.....))))).))))))... ( -41.20) >DroEre_CAF1 17773 91 + 1 UUGUGUUUGACAAAGUUGCGAAACAAGAUGUUAACCCCGGGGGUUGACUCCA---------------UGGUGGAGGCAGGGAUGCAGCAGCAUCUUGAGAAAAGAU-------------- ((((.....))))..........(((((((((..(((((((((....)))).---------------))).))..(((....)))...))))))))).........-------------- ( -25.20) >DroYak_CAF1 18172 91 + 1 UUGUGUUUGACAAAGUUGCGAAACAAGAUGUUAACCCCGAGGGUUGACUCCG---------------AAGUGGAGGCAGGGAUGCAGCAGCAUCUUGAGAAAAGAU-------------- ((((.....)))).(((((...........(((((((...)))))))((((.---------------....))))(((....))).)))))(((((.....)))))-------------- ( -24.40) >DroAna_CAF1 24855 105 + 1 UUGUGUCUGACAAAGUUGCGAAACAAGAUGUUAACCCCGGGGGUUGACUUUU---------------CGUUGGACUUAGGGAUGCAGCAGCAUCUUGAGAAAAGAUCUCGGUGAUCUGUG ....(((..((...(((....)))..((.((((((((...))))))))...)---------------)))..)))...(((((((....)))))))......(((((.....)))))... ( -33.40) >DroPer_CAF1 19282 119 + 1 UUGUGUUUGACAAAGUUGCGCAACAAGAUGUUAACCCCGAGGGUUGACAUCUGUACUCCGUACAGCGUGGUGGA-GUGGGGACGCAACAGCAUCUUGAGAAAAGAUCUCAGAGAUCUCUG ((((((((..((..(..((((....((((((((((((...))))))))))))((((...)))).))))..)...-.)).))))))))(((.(((((((((.....))))).))))..))) ( -42.50) >consensus UUGUGUUUGACAAAGUUGCGAAACAAGAUGUUAACCCCGAGGGUUGACUCCU_______________UGGUGGAGGCAGGGAUGCAGCAGCAUCUUGAGAAAAGAU______________ ((((.....)))).(((((..........((((((((...))))))))...........................(((....))).)))))(((((.....))))).............. (-21.16 = -20.97 + -0.19)



| Location | 8,073,228 – 8,073,319 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.54 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -16.24 |
| Energy contribution | -16.22 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8073228 91 - 27905053 --------------AUCUUUUCUCAAGAUGCUGCUGCAUCCCUGCCUUCAUUC---------------UGGAGUCAACCCCCGGGGUUAACAUCUUGUUUCGCAACUUUGUCAAACACAA --------------............(((((....)))))....((.......---------------.)).((.(((((...))))).))....(((((.(((....))).)))))... ( -17.70) >DroPse_CAF1 19166 119 - 1 CAGAGAUCUCUGAGAUCUUUUCUCAAGAUGCUGUUGCGUCCCCAC-UCCACCACGUUGUACGGAGUACAGAUGUCAACCCUCGGGGUUAACAUCUUGUUGCGCAACUUUGUCAAACACAA .(((((((.....)))))))......((((..(((((((..(.((-(((............)))))..((((((.(((((...))))).)))))).)..)))))))..))))........ ( -36.90) >DroEre_CAF1 17773 91 - 1 --------------AUCUUUUCUCAAGAUGCUGCUGCAUCCCUGCCUCCACCA---------------UGGAGUCAACCCCCGGGGUUAACAUCUUGUUUCGCAACUUUGUCAAACACAA --------------............(((((....)))))...(.((((....---------------.)))).)(((((...))))).......(((((.(((....))).)))))... ( -20.10) >DroYak_CAF1 18172 91 - 1 --------------AUCUUUUCUCAAGAUGCUGCUGCAUCCCUGCCUCCACUU---------------CGGAGUCAACCCUCGGGGUUAACAUCUUGUUUCGCAACUUUGUCAAACACAA --------------............(((((....)))))...(.((((....---------------.)))).)(((((...))))).......(((((.(((....))).)))))... ( -19.80) >DroAna_CAF1 24855 105 - 1 CACAGAUCACCGAGAUCUUUUCUCAAGAUGCUGCUGCAUCCCUAAGUCCAACG---------------AAAAGUCAACCCCCGGGGUUAACAUCUUGUUUCGCAACUUUGUCAGACACAA ...........((((.....))))..(((((....))))).....(((..(((---------------((..((.(((((...))))).))...(((.....))).)))))..))).... ( -23.50) >DroPer_CAF1 19282 119 - 1 CAGAGAUCUCUGAGAUCUUUUCUCAAGAUGCUGUUGCGUCCCCAC-UCCACCACGCUGUACGGAGUACAGAUGUCAACCCUCGGGGUUAACAUCUUGUUGCGCAACUUUGUCAAACACAA .(((((((.....)))))))......((((..(((((((..(.((-(((............)))))..((((((.(((((...))))).)))))).)..)))))))..))))........ ( -36.90) >consensus ______________AUCUUUUCUCAAGAUGCUGCUGCAUCCCUACCUCCACCA_______________AGAAGUCAACCCCCGGGGUUAACAUCUUGUUUCGCAACUUUGUCAAACACAA ..........................(((((....)))))................................((.(((((...))))).))....(((((.(((....))).)))))... (-16.24 = -16.22 + -0.03)

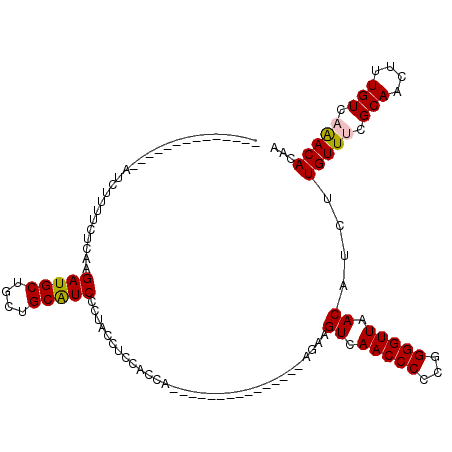

| Location | 8,073,293 – 8,073,388 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 80.16 |
| Mean single sequence MFE | -29.65 |
| Consensus MFE | -18.48 |
| Energy contribution | -18.32 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8073293 95 + 27905053 GAUGCAGCAGCAUCUUGAGAAAAGAU---------------CAGCAGCUGCCCUU-GUCGCAGUGAGAUCACCGAAAACUGACAUUUGACCAUUGGUCAAAGGGUUGACCA-- ...(((((.(((((((.....)))))---------------..)).)))))....-(((...(((....)))........(((.(((((((...)))))))..))))))..-- ( -26.20) >DroPse_CAF1 19245 109 + 1 GACGCAACAGCAUCUUGAGAAAAGAUCUCAGAGAUCUCUGACAGCAGCUGCCCUCCGAAAGAGAGAGAGUUCG----GUUAACAUUUGACCAUUGGUCAAAGGGUUGACCAGA ((.((..(((((((((.....))))).(((((....))))).....))))..((((....).)))...))))(----((((((.(((((((...)))))))..)))))))... ( -36.20) >DroSec_CAF1 19339 95 + 1 GAUGCAGCAGCAUCUUGAGAAAAGAU---------------CAGCAGCUGCCCUU-GUCGCAGUGAGAUCAACGAAAACUGACAUUUGACCACUGGUCAAAGGGUUGACCA-- (((((....))))).........(((---------------(....(((((....-...)))))..))))..........(((.(((((((...)))))))..))).....-- ( -25.20) >DroEre_CAF1 17838 95 + 1 GAUGCAGCAGCAUCUUGAGAAAAGAU---------------CAGCAGCUGCCCUU-GUCGCAGUGAGAUCACCGAGAACUGACAUUUGACCAUUGGUCAAAGGGUUGACCA-- ...(((((.(((((((.....)))))---------------..)).)))))....-(((((.((.((.((.....)).)).)).(((((((...)))))))..).))))..-- ( -26.20) >DroYak_CAF1 18237 95 + 1 GAUGCAGCAGCAUCUUGAGAAAAGAU---------------CAGCAGCUGCCCUU-GUCGCAGUGAGAUCACCGAAAACUGACAUUUGACCAUUGGUCAAAGGGUUGGCCA-- (((((....))))).........(((---------------(....(((((....-...)))))..))))........(..((.(((((((...)))))))..))..)...-- ( -27.90) >DroPer_CAF1 19361 109 + 1 GACGCAACAGCAUCUUGAGAAAAGAUCUCAGAGAUCUCUGACAGCAGCUGCCCUCCGAAAGAGAGAGAGUUCG----GUUAACAUUUGACCAUUGGUCAAAGGGUUGACCAGA ((.((..(((((((((.....))))).(((((....))))).....))))..((((....).)))...))))(----((((((.(((((((...)))))))..)))))))... ( -36.20) >consensus GAUGCAGCAGCAUCUUGAGAAAAGAU_______________CAGCAGCUGCCCUU_GUCGCAGUGAGAUCACCGAAAACUGACAUUUGACCAUUGGUCAAAGGGUUGACCA__ (((((....))))).................................(.((((.........(((....)))............(((((((...))))))))))).)...... (-18.48 = -18.32 + -0.16)



| Location | 8,073,293 – 8,073,388 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.16 |
| Mean single sequence MFE | -30.05 |
| Consensus MFE | -18.86 |
| Energy contribution | -19.14 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.758617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8073293 95 - 27905053 --UGGUCAACCCUUUGACCAAUGGUCAAAUGUCAGUUUUCGGUGAUCUCACUGCGAC-AAGGGCAGCUGCUG---------------AUCUUUUCUCAAGAUGCUGCUGCAUC --........(((((((((...)))))).((((......(((((....))))).)))-))))(((((.((..---------------(((((.....))))))).)))))... ( -28.30) >DroPse_CAF1 19245 109 - 1 UCUGGUCAACCCUUUGACCAAUGGUCAAAUGUUAAC----CGAACUCUCUCUCUUUCGGAGGGCAGCUGCUGUCAGAGAUCUCUGAGAUCUUUUCUCAAGAUGCUGUUGCGUC ...(((.(((..(((((((...))))))).))).))----)((..((((((((.....)))(((((...))))))))))..))(((((.....))))).(((((....))))) ( -34.00) >DroSec_CAF1 19339 95 - 1 --UGGUCAACCCUUUGACCAGUGGUCAAAUGUCAGUUUUCGUUGAUCUCACUGCGAC-AAGGGCAGCUGCUG---------------AUCUUUUCUCAAGAUGCUGCUGCAUC --(((((((....)))))))(.((((((.((........)))))))).)........-....(((((.((..---------------(((((.....))))))).)))))... ( -26.70) >DroEre_CAF1 17838 95 - 1 --UGGUCAACCCUUUGACCAAUGGUCAAAUGUCAGUUCUCGGUGAUCUCACUGCGAC-AAGGGCAGCUGCUG---------------AUCUUUUCUCAAGAUGCUGCUGCAUC --(((((((....)))))))..(((((..((((...((.(((((....))))).)).-...)))).....))---------------))).........(((((....))))) ( -28.60) >DroYak_CAF1 18237 95 - 1 --UGGCCAACCCUUUGACCAAUGGUCAAAUGUCAGUUUUCGGUGAUCUCACUGCGAC-AAGGGCAGCUGCUG---------------AUCUUUUCUCAAGAUGCUGCUGCAUC --...((.....(((((((...)))))))((((......(((((....))))).)))-).))(((((.((..---------------(((((.....))))))).)))))... ( -28.70) >DroPer_CAF1 19361 109 - 1 UCUGGUCAACCCUUUGACCAAUGGUCAAAUGUUAAC----CGAACUCUCUCUCUUUCGGAGGGCAGCUGCUGUCAGAGAUCUCUGAGAUCUUUUCUCAAGAUGCUGUUGCGUC ...(((.(((..(((((((...))))))).))).))----)((..((((((((.....)))(((((...))))))))))..))(((((.....))))).(((((....))))) ( -34.00) >consensus __UGGUCAACCCUUUGACCAAUGGUCAAAUGUCAGUUUUCGGUGAUCUCACUGCGAC_AAGGGCAGCUGCUG_______________AUCUUUUCUCAAGAUGCUGCUGCAUC ..........(((((((((...)))))............(((((....))))).....))))(((((.((.................(((((.....))))))).)))))... (-18.86 = -19.14 + 0.28)

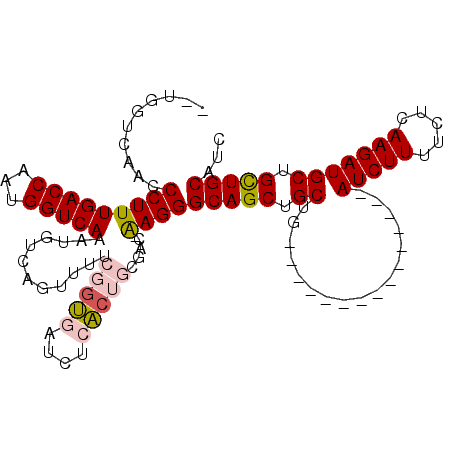
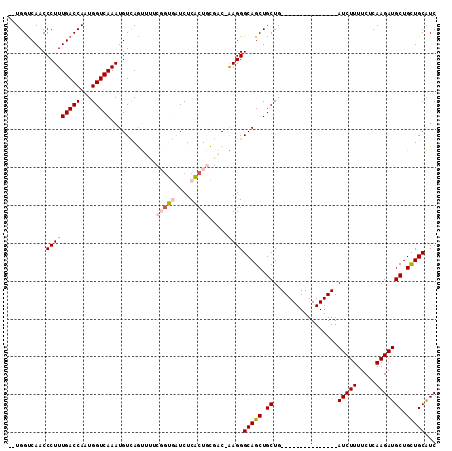
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:23 2006