
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,065,771 – 8,065,941 |
| Length | 170 |
| Max. P | 0.839416 |

| Location | 8,065,771 – 8,065,867 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 82.74 |
| Mean single sequence MFE | -38.20 |
| Consensus MFE | -27.71 |
| Energy contribution | -28.19 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8065771 96 + 27905053 CCAGCCUGGAGCCCAGAGGAGC--GUAGUGGAGCUGAACAUGGAGCUACAGCCCGGAGGAGGACGAAAUCGUCGUUGUGCUCCAGCCAGGUCGGAACU ((.((((((((((((.......--....))).))).....((((((.(((((((...))..((((....))))))))))))))).)))))).)).... ( -38.20) >DroSec_CAF1 11930 86 + 1 CCAGCCUGGAGCACAGGGGAG------------CUGAACAUGGAGCUGCAGCCCGGAGGAGGACGACAUCGUCGUUGUGCUCCAGCCAGGUCGGAACU ((.((((((......(((.((------------((........))))....)))((((.(.((((((...)))))).).))))..)))))).)).... ( -36.50) >DroSim_CAF1 11921 98 + 1 CCAGCCUGGAGCACAGGGGAGCUGGAGUUGGAGCUGAACAUGGAGCUGCUGCCCGGAGGAGGACGACAUCGUCGUUGUGCUCCAGCCAGGUCGGAACU ((.((((((......(((.(((.........((((........))))))).)))((((.(.((((((...)))))).).))))..)))))).)).... ( -41.70) >DroEre_CAF1 10351 78 + 1 CCAGCCUGGAGCCCAGAGGAGC------UGGAGCUGAAGAUGGAGCUGCAGC-CGGAGGAG-------------CUGUGCUCCAGCCAGGUCGGAACU ((.(((((((((((((.....)------))).))).....((((((.(((((-(....).)-------------)))))))))).)))))).)).... ( -39.20) >DroYak_CAF1 10572 86 + 1 CCAGCCUGGAGCCCAGAGGAG------------CUGAAGAUGGAGCUGCAGCCCGGAGGAGGACGACAUCGUCGUGGUGCUCCAGCCAGGUCGGAACU ((.((((((........((.(------------(((.((......)).))))))((((.(..(((((...)))))..).))))..)))))).)).... ( -35.40) >consensus CCAGCCUGGAGCCCAGAGGAGC______UGGAGCUGAACAUGGAGCUGCAGCCCGGAGGAGGACGACAUCGUCGUUGUGCUCCAGCCAGGUCGGAACU ((.((((((....(((.................)))....((((((.(((((.(......)((((....))))))))))))))).)))))).)).... (-27.71 = -28.19 + 0.48)



| Location | 8,065,771 – 8,065,867 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 82.74 |
| Mean single sequence MFE | -35.34 |
| Consensus MFE | -23.50 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.839416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8065771 96 - 27905053 AGUUCCGACCUGGCUGGAGCACAACGACGAUUUCGUCCUCCUCCGGGCUGUAGCUCCAUGUUCAGCUCCACUAC--GCUCCUCUGGGCUCCAGGCUGG ((((..((((..(..(((((.....((((....)))).......((((((.(((.....))))))).)).....--))))).)..)).))..)))).. ( -34.00) >DroSec_CAF1 11930 86 - 1 AGUUCCGACCUGGCUGGAGCACAACGACGAUGUCGUCCUCCUCCGGGCUGCAGCUCCAUGUUCAG------------CUCCCCUGUGCUCCAGGCUGG ........((.(.((((((((((..((((....)))).......((((((.(((.....))))))------------)))...)))))))))).).)) ( -38.10) >DroSim_CAF1 11921 98 - 1 AGUUCCGACCUGGCUGGAGCACAACGACGAUGUCGUCCUCCUCCGGGCAGCAGCUCCAUGUUCAGCUCCAACUCCAGCUCCCCUGUGCUCCAGGCUGG ........((.(.((((((((((..((((....)))).......(((.(((((((........)))).........))).))))))))))))).).)) ( -37.00) >DroEre_CAF1 10351 78 - 1 AGUUCCGACCUGGCUGGAGCACAG-------------CUCCUCCG-GCUGCAGCUCCAUCUUCAGCUCCA------GCUCCUCUGGGCUCCAGGCUGG ....(((.(((((.((((((.(((-------------((.....)-))))..)))))).....(((.(((------(.....)))))))))))).))) ( -33.30) >DroYak_CAF1 10572 86 - 1 AGUUCCGACCUGGCUGGAGCACCACGACGAUGUCGUCCUCCUCCGGGCUGCAGCUCCAUCUUCAG------------CUCCUCUGGGCUCCAGGCUGG ........((.(.(((((((.(((.((((....)))).......((((((.((......)).)))------------)))...)))))))))).).)) ( -34.30) >consensus AGUUCCGACCUGGCUGGAGCACAACGACGAUGUCGUCCUCCUCCGGGCUGCAGCUCCAUGUUCAGCUCCA______GCUCCUCUGGGCUCCAGGCUGG ........((.(.(((((((.....((((....)))).......((((....))))..............................))))))).).)) (-23.50 = -23.90 + 0.40)

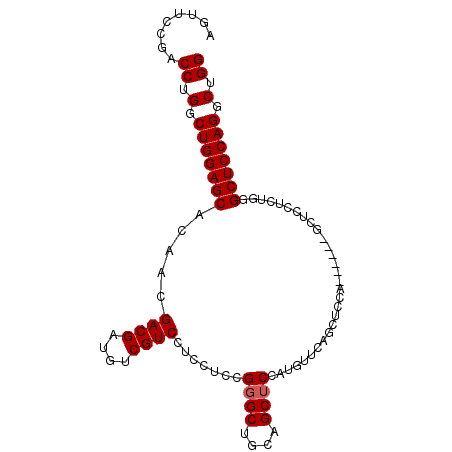
| Location | 8,065,827 – 8,065,941 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.01 |
| Mean single sequence MFE | -37.18 |
| Consensus MFE | -27.38 |
| Energy contribution | -28.43 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8065827 114 + 27905053 GAGGACGAAAUCGUCGUUGUGCUCCAGCCAGGUCGGAACUGAUCUAUGGUAGCCGUUAGUGUCGUCGGCAGUGAAAAGUUGUCGACCUGCACUGGCAAAAU------AUAGUAAAUAGCG ...((((....))))(((((((((((...((((((....)))))).))).))).((((((((.(((((((((....).))))))))..)))))))).....------.......))))). ( -40.70) >DroSec_CAF1 11976 113 + 1 GAGGACGACAUCGUCGUUGUGCUCCAGCCAGGUCGGAACUGAUCUAUGGUAGCCGUUAGUGUCGUCGGCAGUGAAAAGUUGUCGACCUGCACUGGCAG-GU------AUAGUAAAUAGCG ...((((....))))(((((((((((...((((((....)))))).))).))).((((((((.(((((((((....).))))))))..))))))))..-..------.......))))). ( -40.70) >DroSim_CAF1 11979 114 + 1 GAGGACGACAUCGUCGUUGUGCUCCAGCCAGGUCGGAACUGAUCUAUGGUAGCCGUUAGUGUCGUCGGCAGUGAAAAGUUGUCGACCUGCACUGGCAAAAU------AUAGUAAAUAGCG ...((((....))))(((((((((((...((((((....)))))).))).))).((((((((.(((((((((....).))))))))..)))))))).....------.......))))). ( -40.70) >DroEre_CAF1 10402 107 + 1 GAG-------------CUGUGCUCCAGCCAGGUCGGAACUGAUCUAUGGUAGCCGUUAGUGUCGUCGGCAGUGAAAAGUUGUCGACCUGCACUGGCAAAAUACGAGUAUAGUAAAUAGAG ..(-------------((((((((..(((((((((....)))))..))))....((((((((.(((((((((....).))))))))..)))))))).......)))))))))........ ( -39.80) >DroYak_CAF1 10618 114 + 1 GAGGACGACAUCGUCGUGGUGCUCCAGCCAGGUCGGAACUGAUCCAUGGUAGCCGUUAGUGUCGUCGGCAGUGAAAAGUUGUCGACCUGCACUGGCAAAAU------AUAGUAAAUAGAG ...((((....))))..(((...(((....(((((....)))))..)))..)))((((((((.(((((((((....).))))))))..)))))))).....------............. ( -37.00) >DroAna_CAF1 17555 91 + 1 -----CCACUGA------GU-CCUCGGCCAGGUUGAAUCUGAUCUAUGGUGGCCGUUAGUGUCUUCGGCAGUGAAAAGUUGGCGACCUGCUCGUGCAAAAU------AU----------- -----.(((.((------((-....(..(((((...)))))..)...(((.(((((((.((.(....))).))).....)))).))).)))))))......------..----------- ( -24.20) >consensus GAGGACGACAUCGUCGUUGUGCUCCAGCCAGGUCGGAACUGAUCUAUGGUAGCCGUUAGUGUCGUCGGCAGUGAAAAGUUGUCGACCUGCACUGGCAAAAU______AUAGUAAAUAGCG ...((((....))))...(.((((((...((((((....)))))).))).))))((((((((.((((((((.......))))))))..))))))))........................ (-27.38 = -28.43 + 1.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:14 2006