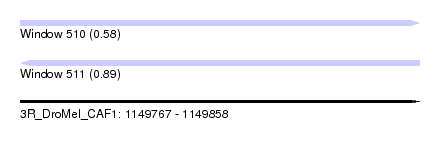
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,149,767 – 1,149,858 |
| Length | 91 |
| Max. P | 0.888524 |

| Location | 1,149,767 – 1,149,858 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 82.75 |
| Mean single sequence MFE | -19.97 |
| Consensus MFE | -14.31 |
| Energy contribution | -14.09 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1149767 91 + 27905053 CAUGUGCUGCAACAAACUCGUUUUUGUUUUUGUUGCAGGCCAAAAACCUAAACCUGCUUGAGUUUGGCUUAUUUCCGUUUAUUUCCACUUC-------- ...(((((((((((((..((....))..))))))))))((((((....(((......)))..)))))).................)))...-------- ( -21.40) >DroSec_CAF1 85265 91 + 1 CAUGUGCUGCAACAAACUCGUUUUUAUUUUUGUUGCAGGCCAAAAACCUGAACCUGCUUGAGCUCGGCUUAUUUCCGUUUAUUUCCACUUC-------- ...(((((((((((((............))))))))))..........(((((.....(((((...))))).....)))))....)))...-------- ( -18.50) >DroSim_CAF1 81128 91 + 1 AAUGUGCUGCAACAAACUCGUUUUUAUUUUUGUUGCAGGCCAAAAACCUGAACCUGCUUGAGCUCGGCUUAUUUCCGUUUAUUUCCACUUC-------- ...(((((((((((((............))))))))))..........(((((.....(((((...))))).....)))))....)))...-------- ( -18.50) >DroEre_CAF1 83738 99 + 1 CAUGUGCUGCAACAAACUCGUUUUUGCUUUUGUUGCAGGCCAAAAACCUGAACCUGCUUGAGCUCGGCUUAUUUCCGUUUAUUUCCACUUCCAAGUGUG ..((..((((((((((..((....))..))))))))))..))......(((((.....(((((...))))).....)))))....((((....)))).. ( -21.40) >DroYak_CAF1 106455 97 + 1 CAUGUGCUGCAACAAACUCGUUUUUG-UUUUGUUGCAGGCUAAAAACCUGAACCUGCUUGGGCUGAGCUUAUUUCCGUUUAUUUCCACUUU-GUGUGUG ...((((((((((((((........)-.))))))))))(((.....((..(......)..))...))).................)))...-....... ( -20.30) >DroAna_CAF1 80459 70 + 1 CAUGUGCUGCAAUAAACUCGUU--------UGUUGCAGGCCAAAAACC------UGCUUA---UUGGCUUAUUUCCGUU----UCCAUGUG-------- ((((((((((((((((....))--------))))))))(((((.((..------...)).---)))))...........----..))))))-------- ( -19.70) >consensus CAUGUGCUGCAACAAACUCGUUUUUGUUUUUGUUGCAGGCCAAAAACCUGAACCUGCUUGAGCUCGGCUUAUUUCCGUUUAUUUCCACUUC________ ...(((((((((((((............))))))))))(((........................))).................)))........... (-14.31 = -14.09 + -0.22)

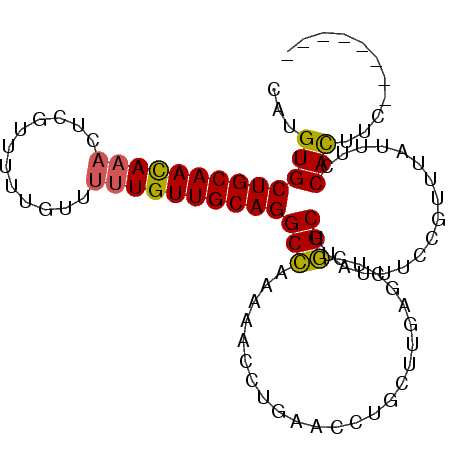
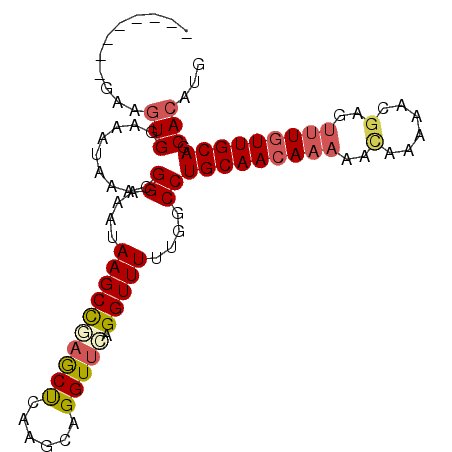
| Location | 1,149,767 – 1,149,858 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 82.75 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -18.53 |
| Energy contribution | -19.30 |
| Covariance contribution | 0.77 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1149767 91 - 27905053 --------GAAGUGGAAAUAAACGGAAAUAAGCCAAACUCAAGCAGGUUUAGGUUUUUGGCCUGCAACAAAAACAAAAACGAGUUUGUUGCAGCACAUG --------...(((.........((..(.((((((((((......))))).))))).)..))(((((((((..(......)..))))))))).)))... ( -23.40) >DroSec_CAF1 85265 91 - 1 --------GAAGUGGAAAUAAACGGAAAUAAGCCGAGCUCAAGCAGGUUCAGGUUUUUGGCCUGCAACAAAAAUAAAAACGAGUUUGUUGCAGCACAUG --------...(((........(((((((.((((..((....)).))))...)))))))..((((((((((............)))))))))))))... ( -24.20) >DroSim_CAF1 81128 91 - 1 --------GAAGUGGAAAUAAACGGAAAUAAGCCGAGCUCAAGCAGGUUCAGGUUUUUGGCCUGCAACAAAAAUAAAAACGAGUUUGUUGCAGCACAUU --------...(((........(((((((.((((..((....)).))))...)))))))..((((((((((............)))))))))))))... ( -24.20) >DroEre_CAF1 83738 99 - 1 CACACUUGGAAGUGGAAAUAAACGGAAAUAAGCCGAGCUCAAGCAGGUUCAGGUUUUUGGCCUGCAACAAAAGCAAAAACGAGUUUGUUGCAGCACAUG ..((((....))))........(((((((.((((..((....)).))))...)))))))((.(((((((((..(......)..)))))))))))..... ( -27.60) >DroYak_CAF1 106455 97 - 1 CACACAC-AAAGUGGAAAUAAACGGAAAUAAGCUCAGCCCAAGCAGGUUCAGGUUUUUAGCCUGCAACAAAA-CAAAAACGAGUUUGUUGCAGCACAUG ..(((..-...)))..........(.(((((((((.((....))..((((((((.....))))).)))....-.......))))))))).)........ ( -20.90) >DroAna_CAF1 80459 70 - 1 --------CACAUGGA----AACGGAAAUAAGCCAA---UAAGCA------GGUUUUUGGCCUGCAACA--------AACGAGUUUAUUGCAGCACAUG --------..((((..----..(((((((..((...---...)).------.)))))))((.(((((.(--------((....))).))))))).)))) ( -17.10) >consensus ________GAAGUGGAAAUAAACGGAAAUAAGCCGAGCUCAAGCAGGUUCAGGUUUUUGGCCUGCAACAAAAACAAAAACGAGUUUGUUGCAGCACAUG ...........(((.........((....((((((((((......))))).)))))....))(((((((((..(......)..))))))))).)))... (-18.53 = -19.30 + 0.77)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:52 2006