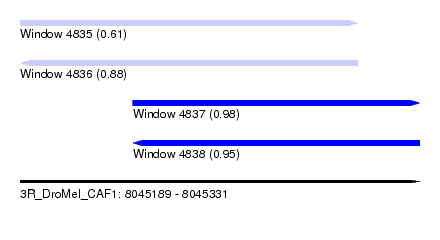
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,045,189 – 8,045,331 |
| Length | 142 |
| Max. P | 0.981985 |

| Location | 8,045,189 – 8,045,309 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -31.99 |
| Consensus MFE | -20.43 |
| Energy contribution | -20.68 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
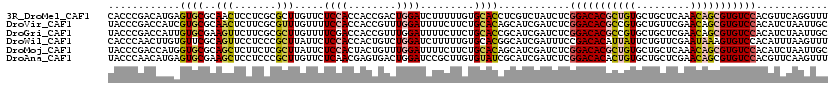
>3R_DroMel_CAF1 8045189 120 + 27905053 CACCCGACAUGAGUGCGCAACUCCUCGCGCUUGUUCUCCACCACCGACUGGAUCUUUUUGUGCACCUCGUCUAUCUCGGACACGCUGUGCUGCUCAAACAGCGUGUCCACGUUCAGGUUU .(((.(((((((((((((((.(((...((...((.....))...))...))).....))))))).))))).......(((((((((((.........)))))))))))..)))..))).. ( -39.90) >DroVir_CAF1 231600 120 + 1 UACCCGACCAUCGUGCGCAACUCUUCGCGUUUGUUUUCCACCACCGUUUGGAUUUUCUUCUGCACAGCAUCGAUCUCGGACACGCCGUGCUGUUCGAACAGCGUGUCCACAUCUAAUUGC ............((((((........))........((((........)))).........)))).(((..(((...((((((((.((.........)).))))))))..)))....))) ( -28.80) >DroGri_CAF1 160733 120 + 1 UACCCGACCAUUGUGCGAAGUUCUUCGCGCUUGUUUUCGACCACCGUUUGGAUUUUCUUCUGCACCGCAUCGAUCUCGGACACGCCGUGCUGCUCGAACAGCGUGUCCACAUCUAAUUGC ....(((..((.(((((((....)))))))..))..))).....((.((((((.......(((...)))........((((((((.((.........)).))))))))..)))))).)). ( -31.70) >DroWil_CAF1 151494 120 + 1 CACCCAACUUGUGUUCGCAGUUCCUCCCGCUUAUUCUCCACCACUGUCUGGAUCUUUUUGUGCACGGCAUCGAUUUCCGACACAUUAUUCUGUUCGAAUAAAGUGUCCACAUUUAAGUUU .....((((((((((((..((.((...(((......((((........)))).......)))...)).))))).....(((((.((((((.....)))))).))))).))))..))))). ( -20.92) >DroMoj_CAF1 180251 120 + 1 UACCCGACCAUGGUGCGCAGCUCUUCUCGCUUAUUCUCCACUACUGUUUGGAUUUUCUUCUGCACAGCAUCGAUCUCGGACACGCUGUGCUGCUCAAACAGCGUGUCCACAUCUAAUUGC .....(((.(((.((.(((((.......))......((((........))))........))).)).))).).))..(((((((((((.........)))))))))))............ ( -33.50) >DroAna_CAF1 119332 120 + 1 UACCCAACAUGAGUGCGAAGCUCCUCCCGCUUGUUCUCAACGAGUGACUGGAUCCGCUUGUGUAUCGCAUCGAUCUCGGACACACUGUGCUGCUCGAACAGCGUGUCCACGUUCAAGUUU ......(((..((((.((...(((...(((((((.....)))))))...)))))))))..)))........((....((((((.((((.........)))).))))))....))...... ( -37.10) >consensus UACCCGACCAGAGUGCGCAGCUCCUCGCGCUUGUUCUCCACCACCGUCUGGAUCUUCUUCUGCACCGCAUCGAUCUCGGACACGCUGUGCUGCUCGAACAGCGUGUCCACAUCUAAGUGC ............((((..(((.......))).....((((........)))).........))))............(((((((((((.........)))))))))))............ (-20.43 = -20.68 + 0.26)


| Location | 8,045,189 – 8,045,309 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -38.90 |
| Consensus MFE | -26.08 |
| Energy contribution | -25.92 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.878915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8045189 120 - 27905053 AAACCUGAACGUGGACACGCUGUUUGAGCAGCACAGCGUGUCCGAGAUAGACGAGGUGCACAAAAAGAUCCAGUCGGUGGUGGAGAACAAGCGCGAGGAGUUGCGCACUCAUGUCGGGUG ..((((((.((((((((((((((.........))))))))))).........(((.(((.(((.....(((..(((((..((.....)).)).)))))).))).))))))))))))))). ( -45.60) >DroVir_CAF1 231600 120 - 1 GCAAUUAGAUGUGGACACGCUGUUCGAACAGCACGGCGUGUCCGAGAUCGAUGCUGUGCAGAAGAAAAUCCAAACGGUGGUGGAAAACAAACGCGAAGAGUUGCGCACGAUGGUCGGGUA (((....(((.((((((((((((.(.....).))))))))))))..)))..)))(((((.........((((........))))........((((....)))))))))........... ( -38.70) >DroGri_CAF1 160733 120 - 1 GCAAUUAGAUGUGGACACGCUGUUCGAGCAGCACGGCGUGUCCGAGAUCGAUGCGGUGCAGAAGAAAAUCCAAACGGUGGUCGAAAACAAGCGCGAAGAACUUCGCACAAUGGUCGGGUA (((....(((.((((((((((((.(.....).))))))))))))..)))..))).((((.((((.....(((.....)))(((..........)))....))))))))............ ( -37.70) >DroWil_CAF1 151494 120 - 1 AAACUUAAAUGUGGACACUUUAUUCGAACAGAAUAAUGUGUCGGAAAUCGAUGCCGUGCACAAAAAGAUCCAGACAGUGGUGGAGAAUAAGCGGGAGGAACUGCGAACACAAGUUGGGUG ...(((...(((((((((.((((((.....)))))).)))))((.........))...))))..)))((((((...(((((.....))..((((......))))...)))...)))))). ( -30.10) >DroMoj_CAF1 180251 120 - 1 GCAAUUAGAUGUGGACACGCUGUUUGAGCAGCACAGCGUGUCCGAGAUCGAUGCUGUGCAGAAGAAAAUCCAAACAGUAGUGGAGAAUAAGCGAGAAGAGCUGCGCACCAUGGUCGGGUA ...........((((((((((((.........)))))))))))).((((.(((.(((((.........((((........)))).....(((.......)))))))).)))))))..... ( -39.50) >DroAna_CAF1 119332 120 - 1 AAACUUGAACGUGGACACGCUGUUCGAGCAGCACAGUGUGUCCGAGAUCGAUGCGAUACACAAGCGGAUCCAGUCACUCGUUGAGAACAAGCGGGAGGAGCUUCGCACUCAUGUUGGGUA ..(((..(.((((((((((((((.(.....).)))))))))))(((((((...))))......((((((((.....((((((.......)))))).)))..))))).)))))))..))). ( -41.80) >consensus AAAAUUAAAUGUGGACACGCUGUUCGAGCAGCACAGCGUGUCCGAGAUCGAUGCGGUGCACAAGAAAAUCCAAACAGUGGUGGAGAACAAGCGCGAAGAGCUGCGCACAAAGGUCGGGUA ...........((((((((((((.(.....).))))))))))))...(((((...((((.........((((........)))).....(((.......)))..))))....)))))... (-26.08 = -25.92 + -0.16)


| Location | 8,045,229 – 8,045,331 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 77.98 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -17.92 |
| Energy contribution | -17.32 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8045229 102 + 27905053 CCACCGACUGGAUCUUUUUGUGCACCUCGUCUAUCUCGGACACGCUGUGCUGCUCAAACAGCGUGUCCACGUUCAGGUUUAACAAAUUGGCCGUCAUUUAG-C .....(((.((.((..(((((..(((((((.......(((((((((((.........))))))))))))))...))))...)))))..)))))))......-. ( -34.91) >DroVir_CAF1 231640 102 + 1 CCACCGUUUGGAUUUUCUUCUGCACAGCAUCGAUCUCGGACACGCCGUGCUGUUCGAACAGCGUGUCCACAUCUAAUUGCAGUAAAUCGGCGCUCAUUUAU-U .....((.(.(((((....(((((.......(....)((((((((.((.........)).)))))))).........))))).))))).).))........-. ( -26.60) >DroGri_CAF1 160773 99 + 1 CCACCGUUUGGAUUUUCUUCUGCACCGCAUCGAUCUCGGACACGCCGUGCUGCUCGAACAGCGUGUCCACAUCUAAUUGCAGUAAAUCGGCGCUCAUUU---- .....((.(.(((((....(((((.......(....)((((((((.((.........)).)))))))).........))))).))))).).))......---- ( -26.60) >DroWil_CAF1 151534 102 + 1 CCACUGUCUGGAUCUUUUUGUGCACGGCAUCGAUUUCCGACACAUUAUUCUGUUCGAAUAAAGUGUCCACAUUUAAGUUUAACAAACUUGCCGUCAUUUUA-C (((.....)))........(((.(((((..........(((((.((((((.....)))))).))))).......(((((.....)))))))))))))....-. ( -23.90) >DroMoj_CAF1 180291 103 + 1 CUACUGUUUGGAUUUUCUUCUGCACAGCAUCGAUCUCGGACACGCUGUGCUGCUCAAACAGCGUGUCCACAUCUAAUUGCAAUAGAUCGGUGCUCAUUGAUUU ....(((.((((......)))).)))((((((((((.(((((((((((.........))))))))))).((......))....)))))))))).......... ( -38.40) >DroPer_CAF1 128458 102 + 1 CCACAGUCUGUAUCUUUUUGUGCACUACAUCGAUCUCGGAUACGCUGUGCUGCUCAAACAGCGUAUCCACAUUUAAAUUCAGUAGAUUUGCCGUCAUUUAU-U .(((((...........)))))..((((...(....)(((((((((((.........))))))))))).............))))................-. ( -24.70) >consensus CCACCGUCUGGAUCUUCUUCUGCACAGCAUCGAUCUCGGACACGCUGUGCUGCUCAAACAGCGUGUCCACAUCUAAUUGCAAUAAAUCGGCCCUCAUUUAU_U ...(((((((..........((.....))........(((((((((((.........)))))))))))..............)))).)))............. (-17.92 = -17.32 + -0.61)



| Location | 8,045,229 – 8,045,331 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 77.98 |
| Mean single sequence MFE | -30.94 |
| Consensus MFE | -19.64 |
| Energy contribution | -18.28 |
| Covariance contribution | -1.35 |
| Combinations/Pair | 1.57 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8045229 102 - 27905053 G-CUAAAUGACGGCCAAUUUGUUAAACCUGAACGUGGACACGCUGUUUGAGCAGCACAGCGUGUCCGAGAUAGACGAGGUGCACAAAAAGAUCCAGUCGGUGG .-.....((((((....(((((...((((.....((((((((((((.........)))))))))))).........))))..))))).....)).)))).... ( -30.34) >DroVir_CAF1 231640 102 - 1 A-AUAAAUGAGCGCCGAUUUACUGCAAUUAGAUGUGGACACGCUGUUCGAACAGCACGGCGUGUCCGAGAUCGAUGCUGUGCAGAAGAAAAUCCAAACGGUGG .-.........(((((.....(((((....(((.((((((((((((.(.....).))))))))))))..))).......)))))..(......)...))))). ( -36.60) >DroGri_CAF1 160773 99 - 1 ----AAAUGAGCGCCGAUUUACUGCAAUUAGAUGUGGACACGCUGUUCGAGCAGCACGGCGUGUCCGAGAUCGAUGCGGUGCAGAAGAAAAUCCAAACGGUGG ----.......(((((...(((((((....(((.((((((((((((.(.....).))))))))))))..)))..)))))))..((......))....))))). ( -38.00) >DroWil_CAF1 151534 102 - 1 G-UAAAAUGACGGCAAGUUUGUUAAACUUAAAUGUGGACACUUUAUUCGAACAGAAUAAUGUGUCGGAAAUCGAUGCCGUGCACAAAAAGAUCCAGACAGUGG .-.....((((((((((((.....))))).......(((((.((((((.....)))))).)))))..........))))).)).........(((.....))) ( -23.50) >DroMoj_CAF1 180291 103 - 1 AAAUCAAUGAGCACCGAUCUAUUGCAAUUAGAUGUGGACACGCUGUUUGAGCAGCACAGCGUGUCCGAGAUCGAUGCUGUGCAGAAGAAAAUCCAAACAGUAG ..............((((((...(((......)))(((((((((((.........))))))))))).)))))).((((((...((......))...)))))). ( -33.70) >DroPer_CAF1 128458 102 - 1 A-AUAAAUGACGGCAAAUCUACUGAAUUUAAAUGUGGAUACGCUGUUUGAGCAGCACAGCGUAUCCGAGAUCGAUGUAGUGCACAAAAAGAUACAGACUGUGG .-......((((((..((((((...........))))))..)))))).......(((((((((((((....)).(((.....)))....))))).).))))). ( -23.50) >consensus A_AUAAAUGACCGCCAAUUUACUGAAAUUAAAUGUGGACACGCUGUUCGAGCAGCACAGCGUGUCCGAGAUCGAUGCGGUGCACAAAAAAAUCCAAACGGUGG ............(((.(((((.......))))).((((((((((((.(.....).))))))))))))...............................))).. (-19.64 = -18.28 + -1.35)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:59 2006