| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,029,535 – 8,029,633 |
| Length | 98 |
| Max. P | 0.947848 |

| Location | 8,029,535 – 8,029,633 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -19.38 |
| Consensus MFE | -11.89 |
| Energy contribution | -13.00 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8029535 98 + 27905053 -GGGG----UGGUGAAGGAA--GGAAAGAAAACUAAAUGCACAUUUAACAAUAAAUGAGCAACGUUUUUCGGAAAUUAACUUUCAACAACAUGUGUUGAAAUGCA -....----...((((((..--.....(((((.....(((.((((((....)))))).)))....))))).........)))))).((((....))))....... ( -15.59) >DroVir_CAF1 214292 103 + 1 -AGAAAGAAAGGAAAAUGGCAGCAAAAGAAAACCAAAUGCGCAUUUAGCAG-AAAUGAGCUGCGUUUUCCGAAAAUUAACUUUCAACAACAUGUGUUGAAAUGCA -.........((((((((.((((....(.....)...(((.......))).-......))))))))))))..........((((((((.....)))))))).... ( -24.20) >DroYak_CAF1 100219 99 + 1 AGGGA----UGGCAAACAGA--CGAAAGAAAACUAAAUGCACAUUUAACAAUAAAUGAGCAACGUUUUUCGGAAAUUAACUUUCAACAACAUGUGUUGAAAUGCA .....----..(((......--(((((((........(((.((((((....)))))).)))...))))))).........((((((((.....))))))))))). ( -20.60) >DroMoj_CAF1 165195 103 + 1 -AGGGAACAAGGAAAAUGGCAGCAAAAGAAAUCCAAAUGCGCACUUAGCAG-AAAUGAGCUGCGUUUUCAGAAAAUUAACUUUCAACAACAUGUGUUGAAAUGCA -.(....)...(((((((.((((....(.....)...(((.......))).-......)))))))))))...........((((((((.....)))))))).... ( -20.90) >DroAna_CAF1 104001 92 + 1 -A---------AUGAAAAUA--GCAAAGAAAACUAAAUGCGCAUUUAACAAUAAAUGAGAAACGUU-UUCGGAAAUUAACUUUCAACAACAUGUGUUGAAACGCA -.---------.........--((...((((((......(.((((((....)))))).)....)))-)))..........((((((((.....)))))))).)). ( -16.90) >DroPer_CAF1 112338 93 + 1 -A-------GUGCGAA-GGA--GAAAAGAAAACUAAAUGCACAUUUAACAAUAAAUGAGCAACGUUUUU-GGAAAUUAACUUUCAACAACAUGUGUUGAAAUGCA -.-------.((((..-...--.....((((((....(((.((((((....)))))).)))..))))))-..........((((((((.....)))))))))))) ( -18.10) >consensus _AGG_____AGGAAAAUGGA__CAAAAGAAAACUAAAUGCACAUUUAACAAUAAAUGAGCAACGUUUUUCGGAAAUUAACUUUCAACAACAUGUGUUGAAAUGCA ...........................((((((....(((.((((((....)))))).)))..))))))...........((((((((.....)))))))).... (-11.89 = -13.00 + 1.11)



| Location | 8,029,535 – 8,029,633 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -20.53 |
| Consensus MFE | -12.24 |
| Energy contribution | -13.38 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
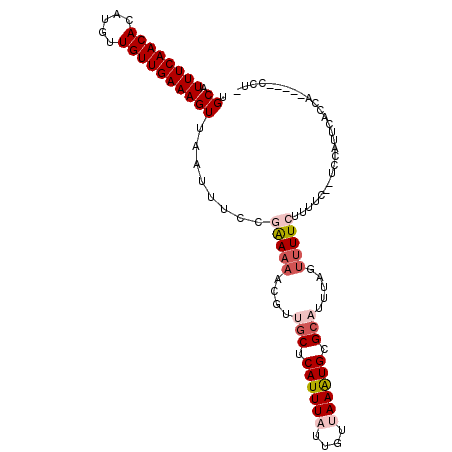
>3R_DroMel_CAF1 8029535 98 - 27905053 UGCAUUUCAACACAUGUUGUUGAAAGUUAAUUUCCGAAAAACGUUGCUCAUUUAUUGUUAAAUGUGCAUUUAGUUUUCUUUCC--UUCCUUCACCA----CCCC- ....((((((((.....))))))))..........(((((....(((.((((((....)))))).))).....))))).....--...........----....- ( -18.40) >DroVir_CAF1 214292 103 - 1 UGCAUUUCAACACAUGUUGUUGAAAGUUAAUUUUCGGAAAACGCAGCUCAUUU-CUGCUAAAUGCGCAUUUGGUUUUCUUUUGCUGCCAUUUUCCUUUCUUUCU- ....((((((((.....))))))))..........((((((.(((((......-..(((((((....)))))))........)))))..)))))).........- ( -26.39) >DroYak_CAF1 100219 99 - 1 UGCAUUUCAACACAUGUUGUUGAAAGUUAAUUUCCGAAAAACGUUGCUCAUUUAUUGUUAAAUGUGCAUUUAGUUUUCUUUCG--UCUGUUUGCCA----UCCCU .(((((((((((.....)))))))).........((((((((..(((.((((((....)))))).)))....)))...)))))--......)))..----..... ( -20.80) >DroMoj_CAF1 165195 103 - 1 UGCAUUUCAACACAUGUUGUUGAAAGUUAAUUUUCUGAAAACGCAGCUCAUUU-CUGCUAAGUGCGCAUUUGGAUUUCUUUUGCUGCCAUUUUCCUUGUUCCCU- .(((((((((((.....))))))))...........(((((.(((((......-...((((((....)))))).........)))))..)))))..))).....- ( -20.97) >DroAna_CAF1 104001 92 - 1 UGCGUUUCAACACAUGUUGUUGAAAGUUAAUUUCCGAA-AACGUUUCUCAUUUAUUGUUAAAUGCGCAUUUAGUUUUCUUUGC--UAUUUUCAU---------U- .(((((((((((.....))))))))..........(((-(((....(.((((((....)))))).)......))))))..)))--.........---------.- ( -16.50) >DroPer_CAF1 112338 93 - 1 UGCAUUUCAACACAUGUUGUUGAAAGUUAAUUUCC-AAAAACGUUGCUCAUUUAUUGUUAAAUGUGCAUUUAGUUUUCUUUUC--UCC-UUCGCAC-------U- (((.((((((((.....))))))))..........-.(((((..(((.((((((....)))))).)))....)))))......--...-...))).-------.- ( -20.10) >consensus UGCAUUUCAACACAUGUUGUUGAAAGUUAAUUUCCGAAAAACGUUGCUCAUUUAUUGUUAAAUGCGCAUUUAGUUUUCUUUUC__UCCAUUCACCA_____CCU_ .((.((((((((.....))))))))))........(((((....(((.((((((....)))))).))).....)))))........................... (-12.24 = -13.38 + 1.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:45 2006