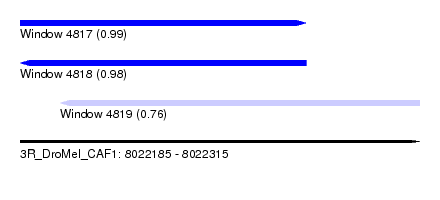
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,022,185 – 8,022,315 |
| Length | 130 |
| Max. P | 0.986135 |

| Location | 8,022,185 – 8,022,278 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 86.62 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -25.97 |
| Energy contribution | -27.33 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.63 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8022185 93 + 27905053 -AACU------------GCACACAGCUGCAGAUGUGCGGCUCGUGUUGCAAUAUCAUUUUUAAUGCAAACCGCAUGCCACAUUAUGCAGCUGCAUUGCCACGUCAC -....------------(((..((((((((...(((.(((..((((((((.((.......)).)))))..)))..))).)))..))))))))...)))........ ( -32.40) >DroPse_CAF1 106889 93 + 1 -AGCU------------GCACCCAGCUGCAGAUGUGCGGCUCGUGUUGCAAUAUCAUUUUUAAUGCAAACCGCAUGCCACAUUAUGCAGCUGCAUUGCCACGUCAC -....------------(((..((((((((...(((.(((..((((((((.((.......)).)))))..)))..))).)))..))))))))...)))........ ( -32.40) >DroGri_CAF1 138164 95 + 1 CA-CUACACCCCCGUUUUCACGCA--CACAGC--AGCGGCUCAUGUUGCA-UAUCAUUUUUAAUGCUAACCACAUGCCGCAUUAUGCAGCUGCAUUGCCAC----- ..-..................(((--....((--((((((....((((((-(..........)))).))).....)))((.....)).)))))..)))...----- ( -21.30) >DroYak_CAF1 92774 93 + 1 -AACU------------GCACACAGCUGCAGAUGUGCGGCUCGUGUUGCAAUAUCAUUUUUAAUGCAAACCGCAUGCCACAUUAUGCAGCUGCAUUGCCACGUCAC -....------------(((..((((((((...(((.(((..((((((((.((.......)).)))))..)))..))).)))..))))))))...)))........ ( -32.40) >DroAna_CAF1 96793 96 + 1 -AACUGC---------UGCACACAGCUGCAGAUGUGCGGCUCGUGUUGCAAUAUCAUUUUUAAUGCAAACCGCAUGCCACAUUAUGCAGCUGUAUUGCCACGUCAC -......---------.(((.(((((((((...(((.(((..((((((((.((.......)).)))))..)))..))).)))..)))))))))..)))........ ( -33.90) >DroPer_CAF1 105115 93 + 1 -AGCU------------GCACCCAGCUGCAGAUGUGCGGCUCGUGUUGCAAUAUCAUUUUUAAUGCAAACCGCAUGCCACAUUAUGCAGCUGCAUUGCCACGUCAC -....------------(((..((((((((...(((.(((..((((((((.((.......)).)))))..)))..))).)))..))))))))...)))........ ( -32.40) >consensus _AACU____________GCACACAGCUGCAGAUGUGCGGCUCGUGUUGCAAUAUCAUUUUUAAUGCAAACCGCAUGCCACAUUAUGCAGCUGCAUUGCCACGUCAC .................(((..((((((((...(((.(((..((((((((.((.......)).)))))..)))..))).)))..))))))))...)))........ (-25.97 = -27.33 + 1.36)



| Location | 8,022,185 – 8,022,278 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 86.62 |
| Mean single sequence MFE | -35.68 |
| Consensus MFE | -30.12 |
| Energy contribution | -30.43 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.46 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8022185 93 - 27905053 GUGACGUGGCAAUGCAGCUGCAUAAUGUGGCAUGCGGUUUGCAUUAAAAAUGAUAUUGCAACACGAGCCGCACAUCUGCAGCUGUGUGC------------AGUU- ..(((...(((.((((((((((...((((((...((..(((((.((.......)).)))))..)).))))))....)))))))))))))------------.)))- ( -36.90) >DroPse_CAF1 106889 93 - 1 GUGACGUGGCAAUGCAGCUGCAUAAUGUGGCAUGCGGUUUGCAUUAAAAAUGAUAUUGCAACACGAGCCGCACAUCUGCAGCUGGGUGC------------AGCU- .....((.(((.(.((((((((...((((((...((..(((((.((.......)).)))))..)).))))))....)))))))).))))------------.)).- ( -35.90) >DroGri_CAF1 138164 95 - 1 -----GUGGCAAUGCAGCUGCAUAAUGCGGCAUGUGGUUAGCAUUAAAAAUGAUA-UGCAACAUGAGCCGCU--GCUGUG--UGCGUGAAAACGGGGGUGUAG-UG -----(((((...((((((((.....))))).))).(((.((((..........)-))))))....))))).--((((..--(.(.((....)).).)..)))-). ( -29.90) >DroYak_CAF1 92774 93 - 1 GUGACGUGGCAAUGCAGCUGCAUAAUGUGGCAUGCGGUUUGCAUUAAAAAUGAUAUUGCAACACGAGCCGCACAUCUGCAGCUGUGUGC------------AGUU- ..(((...(((.((((((((((...((((((...((..(((((.((.......)).)))))..)).))))))....)))))))))))))------------.)))- ( -36.90) >DroAna_CAF1 96793 96 - 1 GUGACGUGGCAAUACAGCUGCAUAAUGUGGCAUGCGGUUUGCAUUAAAAAUGAUAUUGCAACACGAGCCGCACAUCUGCAGCUGUGUGCA---------GCAGUU- .....((.(((.((((((((((...((((((...((..(((((.((.......)).)))))..)).))))))....))))))))))))).---------))....- ( -38.60) >DroPer_CAF1 105115 93 - 1 GUGACGUGGCAAUGCAGCUGCAUAAUGUGGCAUGCGGUUUGCAUUAAAAAUGAUAUUGCAACACGAGCCGCACAUCUGCAGCUGGGUGC------------AGCU- .....((.(((.(.((((((((...((((((...((..(((((.((.......)).)))))..)).))))))....)))))))).))))------------.)).- ( -35.90) >consensus GUGACGUGGCAAUGCAGCUGCAUAAUGUGGCAUGCGGUUUGCAUUAAAAAUGAUAUUGCAACACGAGCCGCACAUCUGCAGCUGUGUGC____________AGUU_ ........(((.((((((((((...((((((...((..(((((.((.......)).)))))..)).))))))....)))))))))))))................. (-30.12 = -30.43 + 0.31)


| Location | 8,022,198 – 8,022,315 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.48 |
| Mean single sequence MFE | -41.25 |
| Consensus MFE | -28.94 |
| Energy contribution | -28.80 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8022198 117 - 27905053 ---AAAGCGGAAGGCGGAUGGGGAUGGAGAAACCGCAGCGGUGACGUGGCAAUGCAGCUGCAUAAUGUGGCAUGCGGUUUGCAUUAAAAAUGAUAUUGCAACACGAGCCGCACAUCUGCA ---....(....)(((((((((..........))...(((((..((((((((((((((..(.....)..))((((.....))))......)).))))))..)))).))))).))))))). ( -42.20) >DroPse_CAF1 106902 104 - 1 ----------------UAGGGCGCAGGGGCCACCACAGCGGUGACGUGGCAAUGCAGCUGCAUAAUGUGGCAUGCGGUUUGCAUUAAAAAUGAUAUUGCAACACGAGCCGCACAUCUGCA ----------------......((((((....))...(((((..((((((((((((((..(.....)..))((((.....))))......)).))))))..)))).)))))....)))). ( -37.00) >DroEre_CAF1 91300 118 - 1 GGAGA--GGAGAGCCGGAUGGGGCGGUGGAAAUCGCAGCGGUGACGUGGCAAUGCAGCUGCAUAAUGUGGCAUGCGGUUUGCAUUAAAAAUGAUAUUGCAACACGAGCCGCACAUCUGCA .....--.....((.(((((..(((((....))))).(((((..((((((((((((((..(.....)..))((((.....))))......)).))))))..)))).))))).))))))). ( -44.30) >DroYak_CAF1 92787 114 - 1 GGGGAAGGGGAAGGCGGAUGGG------AAAACCGCAGCGGUGACGUGGCAAUGCAGCUGCAUAAUGUGGCAUGCGGUUUGCAUUAAAAAUGAUAUUGCAACACGAGCCGCACAUCUGCA .............(((((((((------....))...(((((..((((((((((((((..(.....)..))((((.....))))......)).))))))..)))).))))).))))))). ( -40.70) >DroAna_CAF1 96809 120 - 1 GGGGAGGCGGCGGUGAGAGAGCGGCGGGAAAACCGCAGCGGUGACGUGGCAAUACAGCUGCAUAAUGUGGCAUGCGGUUUGCAUUAAAAAUGAUAUUGCAACACGAGCCGCACAUCUGCA (.(((.(((((.(((........((((.....)))).(((....))).((((((((((..(.....)..))((((.....))))......)).))))))..)))..)))))...))).). ( -46.30) >DroPer_CAF1 105128 104 - 1 ----------------UAGGGCGCAGGGGCCACCACAGCGGUGACGUGGCAAUGCAGCUGCAUAAUGUGGCAUGCGGUUUGCAUUAAAAAUGAUAUUGCAACACGAGCCGCACAUCUGCA ----------------......((((((....))...(((((..((((((((((((((..(.....)..))((((.....))))......)).))))))..)))).)))))....)))). ( -37.00) >consensus ___GA___GG_AG_CGGAGGGCGCAGGGGAAACCGCAGCGGUGACGUGGCAAUGCAGCUGCAUAAUGUGGCAUGCGGUUUGCAUUAAAAAUGAUAUUGCAACACGAGCCGCACAUCUGCA .....................................(((((..((((((((((((((..(.....)..))((((.....))))......)).))))))..)))).)))))......... (-28.94 = -28.80 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:41 2006