
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,993,675 – 7,993,782 |
| Length | 107 |
| Max. P | 0.918818 |

| Location | 7,993,675 – 7,993,782 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.31 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -18.98 |
| Energy contribution | -18.23 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7993675 107 + 27905053 GG-----AGCCGCUUCCGGGUGUAUUUUGCUAAAUUGCAAAUAUUUCUCAAUUCGCAUGCAUCAGCAGAGAAUGCAAUGCAAACAAUUUUCACCCAGCGGCCACAAUA-------AAAA- (.-----.((((((...(((((...(((((......))))).............(((((((((......).)))).))))..........)))))))))))..)....-------....- ( -32.70) >DroVir_CAF1 148537 116 + 1 GGCUUUUGAACGCUUCCGGGCGCAUUUUGCUAAAUUGCAAAUAUUUUCCAAUUCGCAUGCAUCAGCAGC--CCAAAAUGCAAACAAUUUU-GCCCAGCAGCCACAAUGGGCCACAAAAA- ((((((((...(((.(.((((((((((((...(((((.((.....)).))))).((.(((....)))))--.))))))))(((....)))-)))).).)))..))).))))).......- ( -29.60) >DroGri_CAF1 96975 116 + 1 UGGCAUCGAAUGCUUCCGGGCGUAUUUUGCUAAAUUGCAAAUAUUUUCCAAUUCGCAUGCAUCAGCAGC--ACAAUAUGCAAACAAUUUU-GCCCAGCGGCCACAAUGGGCCACAAAAA- .(((((((........)))(((((((.((((....(((.(((........))).))).((....)))))--).))))))).........)-)))..(.((((......)))).).....- ( -29.60) >DroWil_CAF1 90245 105 + 1 -G-----AAAUGCUUCCGGGCGCAUUUUGCUAAAUUGCAAAUAUUUUUCAAUUCGCAUGCAUCAGCA-ACAUUGAAAUGCAAACAAUUUU-GCCCAGCAGCCACAGCA-------ACAAC -.-----...((((....(((((..(((((......)))))....(((((((..((........)).-..))))))).(((((....)))-))...)).)))..))))-------..... ( -23.00) >DroMoj_CAF1 105580 116 + 1 AGGUUUUGCACGCUUCCGGGCGCAUUUUGCUAAAUUGCAAAUAUUUUCCAAUUCGCAUGCAUCAGCAGC--ACGAAAUGCAAACAAUUUU-GCCUGGCAGCCACAAUGGCUCACAAAAA- ...(((((.......(((((((((((((((......))................((.(((....)))))--..)))))))(((....)))-)))))).((((.....))))..))))).- ( -28.90) >DroAna_CAF1 67633 107 + 1 GG-----GACCGCUUCCGGGUGUAUUUUGCUAAAUUGCAAAUAUUUCUCGAAUCGCAUGCAUCAGCAGAGAAUGCAAUGCAAACAAUUUUCGCCCAGCGGCCACAAUA-------AAAA- .(-----(.(((((...(((((...(((((...((((((.....((((((.....).(((....))))))))))))))))))).......))))))))))))......-------....- ( -31.10) >consensus GG_____GAACGCUUCCGGGCGCAUUUUGCUAAAUUGCAAAUAUUUUCCAAUUCGCAUGCAUCAGCAGA__ACGAAAUGCAAACAAUUUU_GCCCAGCAGCCACAAUA_______AAAA_ ...........(((.(.((((....(((((......))))).............((((((....)))..........)))...........)))).).)))................... (-18.98 = -18.23 + -0.75)
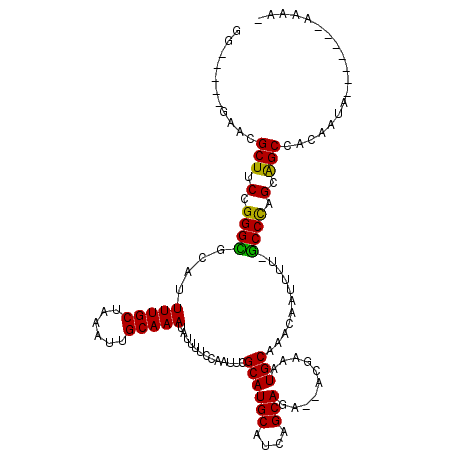
| Location | 7,993,675 – 7,993,782 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.31 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -23.41 |
| Energy contribution | -23.55 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
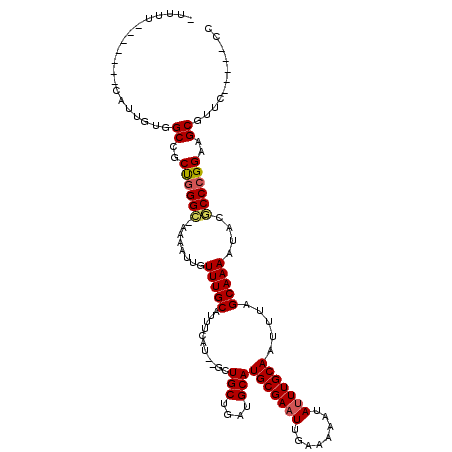
>3R_DroMel_CAF1 7993675 107 - 27905053 -UUUU-------UAUUGUGGCCGCUGGGUGAAAAUUGUUUGCAUUGCAUUCUCUGCUGAUGCAUGCGAAUUGAGAAAUAUUUGCAAUUUAGCAAAAUACACCCGGAAGCGGCU-----CC -....-------......((((((((((((....((((.(((((((((.....))).)))))).))))...........(((((......)))))...)))))...)))))))-----.. ( -36.60) >DroVir_CAF1 148537 116 - 1 -UUUUUGUGGCCCAUUGUGGCUGCUGGGC-AAAAUUGUUUGCAUUUUGG--GCUGCUGAUGCAUGCGAAUUGGAAAAUAUUUGCAAUUUAGCAAAAUGCGCCCGGAAGCGUUCAAAAGCC -.....(..(((......)))..)..(((-....(((..(((.((((((--((((((((....(((((((........)))))))..))))))......)))))))))))..)))..))) ( -37.60) >DroGri_CAF1 96975 116 - 1 -UUUUUGUGGCCCAUUGUGGCCGCUGGGC-AAAAUUGUUUGCAUAUUGU--GCUGCUGAUGCAUGCGAAUUGGAAAAUAUUUGCAAUUUAGCAAAAUACGCCCGGAAGCAUUCGAUGCCA -......((((..((((..((..((((((-..........((((...))--))((((((....(((((((........)))))))..))))))......))))))..))...)))))))) ( -36.00) >DroWil_CAF1 90245 105 - 1 GUUGU-------UGCUGUGGCUGCUGGGC-AAAAUUGUUUGCAUUUCAAUGU-UGCUGAUGCAUGCGAAUUGAAAAAUAUUUGCAAUUUAGCAAAAUGCGCCCGGAAGCAUUU-----C- .....-------.......(((.((((((-.(((((((.((..(((((((.(-(((........)))))))))))..))...))))))).((.....)))))))).)))....-----.- ( -31.10) >DroMoj_CAF1 105580 116 - 1 -UUUUUGUGAGCCAUUGUGGCUGCCAGGC-AAAAUUGUUUGCAUUUCGU--GCUGCUGAUGCAUGCGAAUUGGAAAAUAUUUGCAAUUUAGCAAAAUGCGCCCGGAAGCGUGCAAAACCU -.((((((.((((.....)))).....((-(((....(((.((.(((((--(.(((....))))))))).)).)))...)))))......))))))(((((.(....).)))))...... ( -35.30) >DroAna_CAF1 67633 107 - 1 -UUUU-------UAUUGUGGCCGCUGGGCGAAAAUUGUUUGCAUUGCAUUCUCUGCUGAUGCAUGCGAUUCGAGAAAUAUUUGCAAUUUAGCAAAAUACACCCGGAAGCGGUC-----CC -....-------......(((((((((((((..(((((.(((((((((.....))).)))))).)))))))).......(((((......))))).....)))...)))))))-----.. ( -33.90) >consensus _UUUU_______CAUUGUGGCCGCUGGGC_AAAAUUGUUUGCAUUUCAU__GCUGCUGAUGCAUGCGAAUUGAAAAAUAUUUGCAAUUUAGCAAAAUACGCCCGGAAGCGUUC_____CC ...................((..((((((........(((((...........(((....)))(((((((........))))))).....)))))....))))))..))........... (-23.41 = -23.55 + 0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:28 2006